PLD1 mutations - reading the types of mutations
7 views
Skip to first unread message
Zenovia Gonzalez
Nov 29, 2022, 11:36:05 PM11/29/22
to cbiop...@googlegroups.com
Good evening!
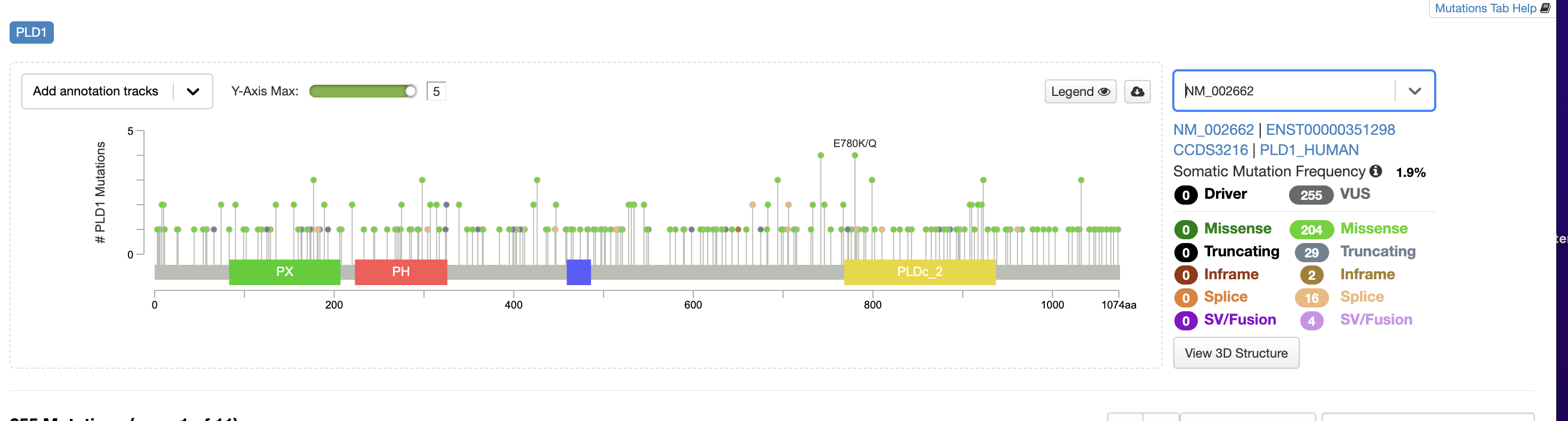
I’m currently looking at PLD1 mutations in the PanCancer Atlas. I’m curious to know how to read different types of mutations. Specifically truncated, inframe, splice, and SV mutations? I attached a screenshot below. I appreciate your help!

Nikolaus Schultz
Nov 29, 2022, 11:43:19 PM11/29/22
to Zenovia Gonzalez, cbiop...@googlegroups.com
Hi Zenovia,
We categorize all nonsense and frameshift mutations as truncating. Splice mutations are those that are predicted to alter the splicing of a gene, and SVs are structural variants, typically larger rearrangements on the genomic level, which sometimes lead to gene fusions.
I hope this is helpful.
Keep in mind that all variants observed in this gene and variants of unknown significance, meaning there is no evidence yet that any of them contribute to tumor development, spread, or sensitivity to therapy.
Niki.
On Nov 29, 2022, at 11:35 PM, Zenovia Gonzalez <zgonza...@gmail.com> wrote:
Good evening!I’m currently looking at PLD1 mutations in the PanCancer Atlas. I’m curious to know how to read different types of mutations. Specifically truncated, inframe, splice, and SV mutations? I attached a screenshot below. I appreciate your help!
<Screen Shot 2022-11-29 at 11.32.08 PM.png>
--
You received this message because you are subscribed to the Google Groups "cBioPortal for Cancer Genomics Discussion Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cbioportal+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cbioportal/CANaGvWaVNwscm_EmC1XzqT5qt-7yDYVquOvzWygU_WpLebiu_w%40mail.gmail.com.
Reply all
Reply to author
Forward
0 new messages
