Somatic mutations for cancer types
7 views
Skip to first unread message
prasun chakraborty
Mar 21, 2023, 1:57:26 PM3/21/23
to cbiop...@googlegroups.com
Good Evening,
I want to look for individual cancers and find out the somatic mutations for the genes which are affected and list them according to their occurrence.
How will I use the database ? I mean I want to rank genes for example lung cancer , for which gene X has more than 1% somatic mutation clinically tested. Then make a panel with genes to study lung cancer.
Best regards,
Prasun.
Benjamin Gross
Mar 21, 2023, 9:12:34 PM3/21/23
to prasun chakraborty, cbiop...@googlegroups.com
Hi Prasun,
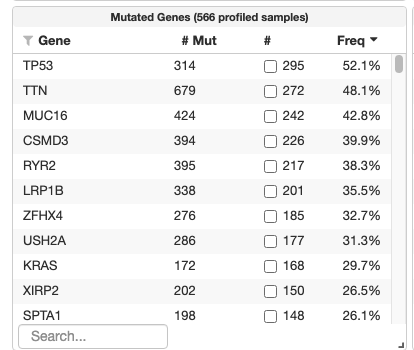
One place to start is by visiting cbioportal.org and selecting one or more lung cancer studies and selecting the “Explore Selected Studies” button. This will take you to the study view where you can see the mutated genes - frequency table across the cohort (see below).
I suggest you visit the “Tutorials/Webinars” page (selection at the header of all pages) to learn more about how to use the cBioPortal.
Regards,
Benjamin


--
You received this message because you are subscribed to the Google Groups "cBioPortal for Cancer Genomics Discussion Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cbioportal+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cbioportal/CAEBDj4a1hVhaqFh_nCcKpjw19tjCquEzHtM9k3ahmjKaCaq_YA%40mail.gmail.com.
Reply all
Reply to author
Forward
0 new messages
