Memory issues with captain proto structs?
53 views
Skip to first unread message
Brando Miranda
Mar 30, 2021, 1:56:53 PM3/30/21
to Cap'n Proto
Hi,
file_name = self.convert_to_local_home_path(file_name)
f = open(file_name)
...
I am doing machine learning with captain proto (because captain proto is good at communicating between python and different languages).
The challenge I have is that my data is represented in captain proto structs. I load a batch of these proto structs every so often to process them with a Neural Network. However, eventually after a certain number of iterations it seems I allocated all the system's memory and I get a SIGKILL from OOM.
I am unsure why this would happen or where (I've been memory profiling my code all day but it is difficult to figure out what part is breaking). I am fairly sure it has to do with captain proto because I used to have a version of the data set with json files and I didn't have this error but now I do. I could directly use the captain proto dataset to create a new json file data set to really figure out if that is the case but it seems redundant.
I thought it could be that I open a captain proto struct and then I close it:
file_name = self.convert_to_local_home_path(file_name)
f = open(file_name)
...
bunch of processing to make it into my python class
....
f.close()
return x, y
I am explicitly closing the file from captain proto so I'd assume that isn't the case. But anyway, is there a way to really check if the memory errors I am getting are due to captain proto or not?
Thanks, Brando
Kenton Varda
Mar 31, 2021, 11:56:16 AM3/31/21
to Brando Miranda, Cap'n Proto
Hi Brando,
It's hard for us to guess what might be the problem without seeing more code.
-Kenton
--
You received this message because you are subscribed to the Google Groups "Cap'n Proto" group.
To unsubscribe from this group and stop receiving emails from it, send an email to capnproto+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/capnproto/ac8e4eba-11e9-44cc-9095-4313e4b7e544n%40googlegroups.com.
Brando Miranda
Mar 31, 2021, 12:19:07 PM3/31/21
to Kenton Varda, Cap'n Proto
Hi Kenton,
Thanks for the reply. I didn't want to overwhelm you guys, its been hard to decide what to share.
Perhaps this will be a good peak into the main function giving me problems:
class DagDataset(Dataset):
def __init__(self, path2dataprep, path2hash2idx, split):
self.split = split
self.path2dataprep = path2dataprep
db = torch.load(self.path2dataprep)
self.data_prep = db['data_prep']
self.list_files_current_split = self.data_prep.flatten_lst_files_split[self.split]
self.list_counts_current_split = self.data_prep.counts[self.split]
self.list_cummulative_sum_current_split = self.data_prep.cummulative_sum[self.split]
self.list_cummulative_end_index_current_split = self.data_prep.cummulative_end_index[self.split]
self.length = sum(self.list_counts_current_split)
#
self.path2hash2idx = path2hash2idx
db = torch.load(self.path2hash2idx)
self.hash2idx = db['hash2idx']
def __len__(self):
return self.length
def __getitem__(self, idx: int) -> DagNode:
# gets the file idx for the value we want
file_idx = bisect.bisect_left(self.list_cummulative_end_index_current_split, idx)
# now get the actual file name
file_name = self.list_files_current_split[file_idx]
# get the file with proof steps
file_name = self.convert_to_local_home_path(file_name)
f = open(file_name)
current_dag_file = dag_api_capnp.Dag.read_packed(f, traversal_limit_in_words=2 ** 64 - 1)
# current_dag_file = dag_api_capnp.Dag.read_packed(f)
# - global idx 2 idx relative to this file
prev_cummulative_sum = self.get_previous_cummulative_sum(file_idx)
idx_rel_this_file = idx - prev_cummulative_sum
# - data point
node_idx = current_dag_file.proofSteps[idx_rel_this_file].node
tactic_hash = current_dag_file.proofSteps[idx_rel_this_file].tactic
tactic_label = self.hash2idx[tactic_hash]
# - get Node obj
node_ref = NodeRef(node_idx, 0) # indicates it's in the current file this cased named current_dag_file
node = DagNode(current_dag_file, node_ref)
# node = current_dag_file
f.close()
return node, tactic_label
my suspicion is that its the this line of code that is giving me issues.:
current_dag_file = dag_api_capnp.Dag.read_packed(f, traversal_limit_in_words=2 ** 64 - 1)
I think if I return the current_dag_file reference directly and do a del and gc.collect() instead of the wrapper around it that I defined called DagNode, it seems to solve the memory problem (perhaps though I am not 100% but fairly sure).
I will confirm if this is true in a sec.
Brando Miranda
Mar 31, 2021, 12:20:49 PM3/31/21
to Kenton Varda, Cap'n Proto
Btw, this is the code getting the data each iteration:
# @profile
def train(self, n_epoch):
import time
self.tactic_predictor.train()
avg_loss = AverageMeter('train loss')
avg_acc = AverageMeter('train accuracy')
# iterations = len(self.dataloaders['train'])
# bar = ProgressBar(max_value=iterations)
self.dataloaders['train'] = iter(self.dataloaders['train'])
# for i, data_batch in enumerate(self.dataloaders['train']):
for i in range(len(self.dataloaders['train'])):
data_batch = next(self.dataloaders['train'])
data_batch = process_batch_ddp(self.opts, data_batch)
# loss, logits = self.tactic_predictor(data_batch)
# acc = accuracy(output=logits, target=data_batch[1])
# avg_loss.update(loss, self.opts.batch_size)
# avg_acc.update(acc, self.opts.batch_size)
self.log(f'{i=}')
#self.log(f"{i=}: {loss=}")
# self.optimizer.zero_grad()
# loss.backward() # each process synchronizes it's gradients in the backward pass
# self.optimizer.step() # the right update is done since all procs have the right synced grads
# del loss
# del logits
# del data_batch
gc.collect()
# bar.update(i)
if i >= 10:
time.sleep(2)
sys.exit()
return avg_loss.item(), avg_acc.item()
Brando Miranda
Mar 31, 2021, 12:25:39 PM3/31/21
to Kenton Varda, Cap'n Proto
Hmmm it seems it takes less effort to remove captain proto files directly compared to removing my wrapper class
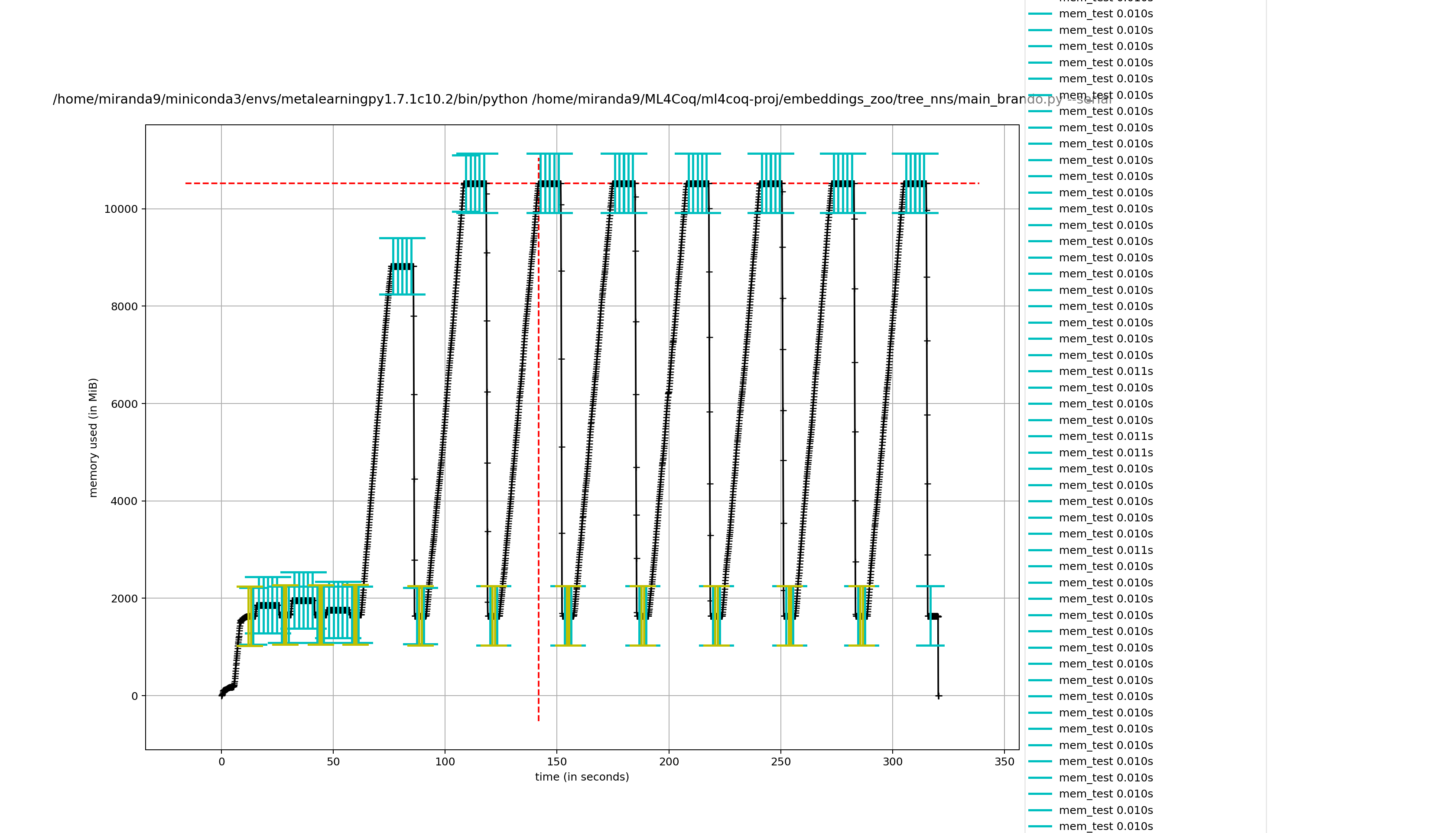
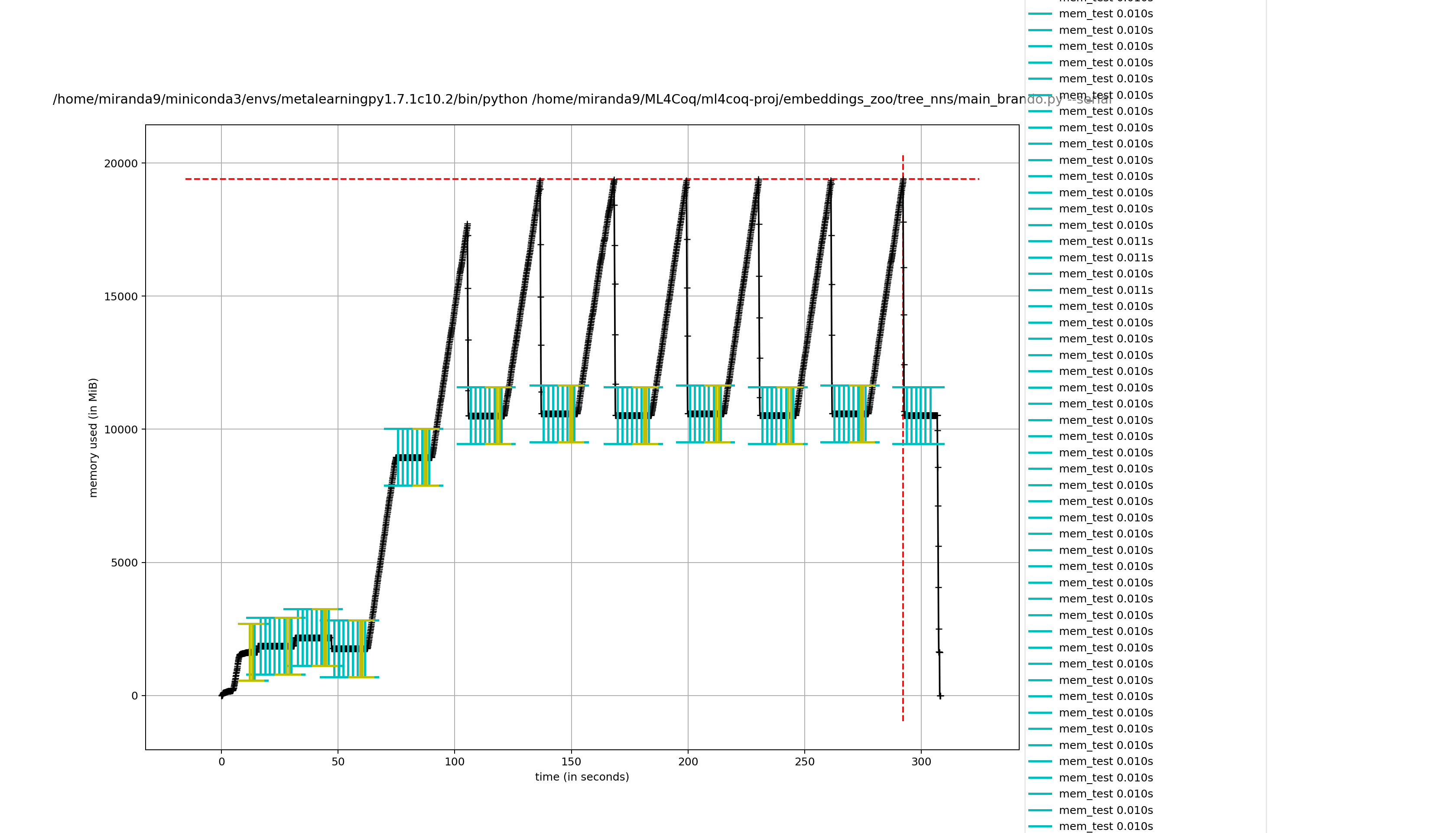
removing captain proto directly

not removing it directly (removing wrapper class)

Brando Miranda
Mar 31, 2021, 6:21:00 PM3/31/21
to Kenton Varda, Cap'n Proto
Hi Kenton,
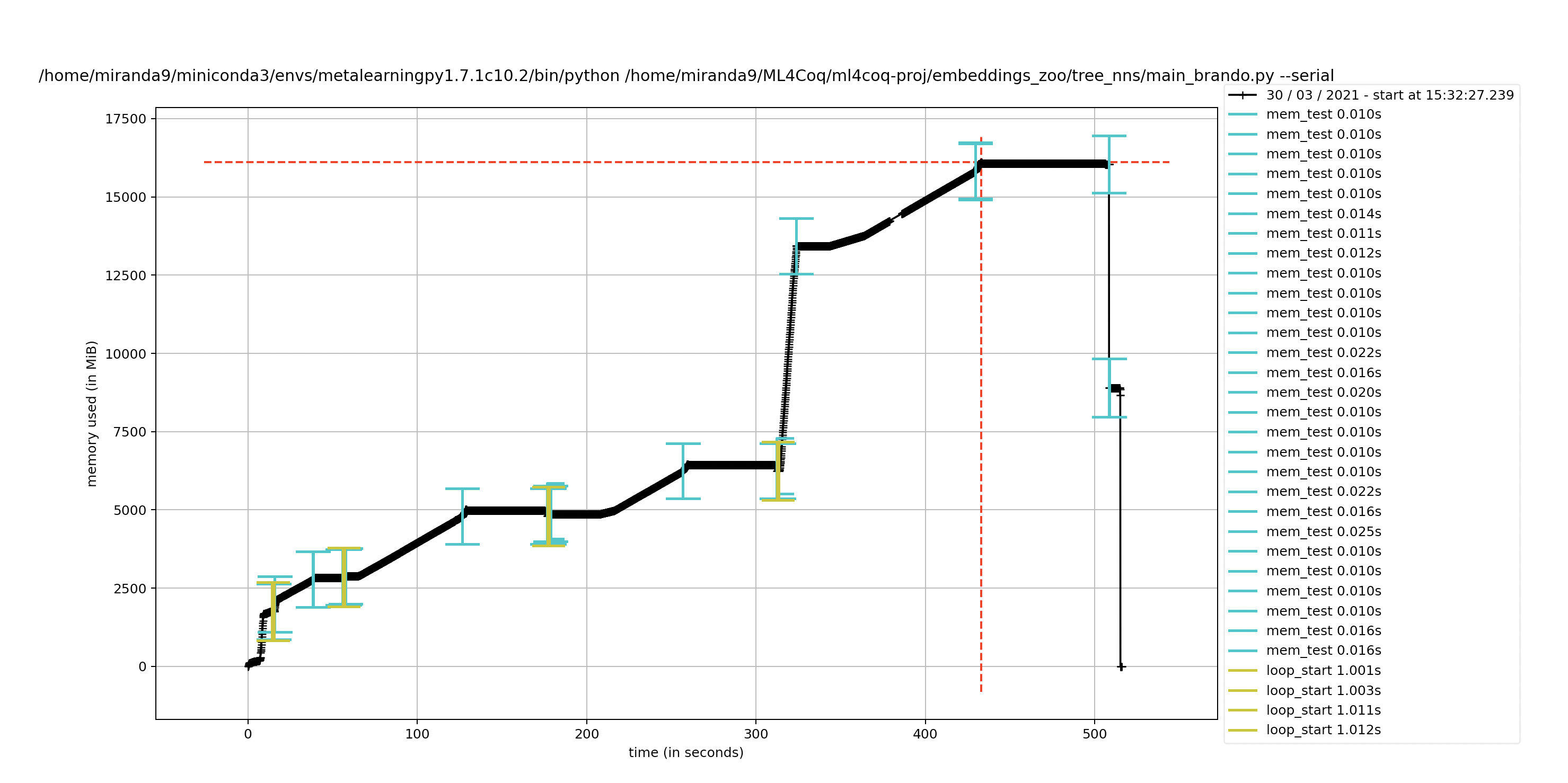
I was wondering, I have dags encoded in captain proto structs. I thought they were the same as my json representations but I am having issues with the captain proto ones. Do you think captain proto might be leaving open files somewhere if I traverse them with a neural network?
the plot with the neural net gives issues even though I explicitly del all the outputs of them every loop

# time.sleep(1)
loss, logits = self.tactic_predictor(data_batch)
# time.sleep(1)
# self.mem_test()
# time.sleep(1)
# acc = accuracy(output=logits, target=data_batch[1])
# avg_loss.update(loss, self.opts.batch_size)
# avg_acc.update(acc, self.opts.batch_size)
self.log(f'{i=}')
self.log(f"{i=}: {loss=}")
# time.sleep(1)
# self.mem_test()
# time.sleep(1)
# self.optimizer.zero_grad()
# loss.backward() # each process synchronizes it's gradients in the backward pass
# self.optimizer.step() # the right update is done since all procs have the right synced grads
# time.sleep(1)
# self.mem_test()
# time.sleep(1)
del loss
del logits
del data_batch
gc.collect()
Kenton Varda
Mar 31, 2021, 8:13:51 PM3/31/21
to Brando Miranda, Cap'n Proto
Hi Brando,
Since you're code is quite complicated, it's hard to tell whether the bug might be in your code vs. Cap'n Proto. If you believe it's a bug in Cap'n Proto, then the best thing to do would be to narrow it down into a minimal self-contained test case. That is, reduce your code to a very short program that you believe clearly shouldn't OOM, but does. Then give that to us so we can run it and debug it ourselves.
-Kenton
Brando Miranda
Apr 2, 2021, 2:13:35 PM4/2/21
to Kenton Varda, Cap'n Proto
Hi Kenton,
Thanks for your kind reply. I agree, it's really hard to help me debug it without providing more code. I have provided small codes multiple times to help track bugs in libraries (e.g. pytorch) and I'd be more than happy to do it here if I knew where to start. Perhaps, while I keep debugging and figuring out what part to reproduce I can ask a leading question that could help me.
When one open as a captain proto file with the `read_packed` (e.g. current_dag_file = dag_api_capnp.Dag.read_packed(f, traversal_limit_in_words=2 ** 64 - 1) do I need to explicitly close this captain proto _StructModule? for example I saw the libraries `finish()` method and was wondering if I had to use that or something else. I am definitively closing the python file at some point (before I return the captain proto struct actually) but I was wondering if you thought I had to close it somewhere else.
Thanks for your time, it's appreciated.
Sincerely, Brando
Kenton Varda
Apr 4, 2021, 9:37:09 PM4/4/21
to Brando Miranda, Cap'n Proto
Hi Brando,
To be honest, I don't know much about pycapnp -- I wrote the main C++ implementation, which pycapnp uses, but I'm not that familiar with pycapnp itself.
I saw this issue was just filed, maybe it is related to your problem: https://github.com/capnproto/pycapnp/issues/245
-Kenton
Brando Miranda
Apr 28, 2021, 3:47:47 PM4/28/21
to Kenton Varda, Cap'n Proto
Thanks! I will check it out. I posted a SO question for future reference and help for others:
Thanks for your time Kenton
Reply all
Reply to author
Forward
0 new messages
