BirdVoxDetect not recogizing SAVS or GCTH
Kevin Tolan
Wim van Dam
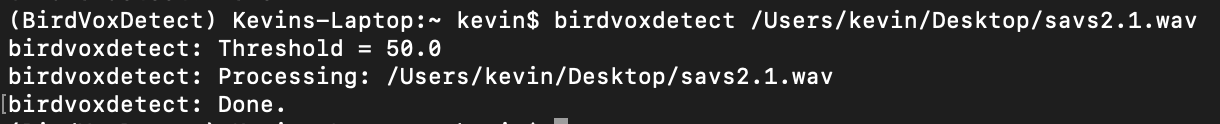
birdvoxdetect --threshold 20 savs2.1.wav
--
You received this message because you are subscribed to the Google Groups "birdvox" group.
To unsubscribe from this group and stop receiving emails from it, send an email to birdvox+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/birdvox/1b49d972-f2c5-4512-bfba-259639f1f0aen%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Vincent Lostanlen
Hello Kevin, Wim, and all,
I agree with Wim, the most probable cause is that the default value of the detection threshold (=50) is too high.
How long is the file savs2.1.wav ? BirdVoxDetect does not work well on very short files because it cannot estimate background noise properly.
I'd be curious to receive the file privately if you're OK with
sharing it. This way i'll be able to investigate further.
Sincerely,
Vincent.
To view this discussion on the web visit https://groups.google.com/d/msgid/birdvox/CAEYyNyRSc9SnzVSCvXcb2vurUAeqYF0gwkPGgQej%3D-8NndZ8kA%40mail.gmail.com.
Kevin Tolan
You received this message because you are subscribed to a topic in the Google Groups "birdvox" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/birdvox/DeqqYAZ-nhc/unsubscribe.
To unsubscribe from this group and all its topics, send an email to birdvox+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/birdvox/f845f4fc-9d0f-aec0-50a0-64e2794dde59%40nyu.edu.
Vincent Lostanlen
Hello everyone,
Following my earlier discussion with Kevin Tolan et al., here is
a (hopefully) useful tip on how to query BirdVoxDetect not at the
species level but at the family level (American sparrow / Cardinal
/ Thrush/ Warbler / other).
"Thank you for the file. I have re-run BVD (BirdVoxDetect) with a threshold of 30 and the flight call is correctly detected.
Furthermore, i passed --predict-proba as an optional argument to BVD in order to obtain a comprehensive output of the taxonomical neural network classifier in JSON format.
In Python:
filepath = "savs2.1.wav"
df = birdvoxdetect.process_file(
filepath,
output_dir="kevintolan",
export_clips=True,
export_confidence=True,
export_logger=True,
predict_proba=True,
threshold=30.0)
And here are the species for the flight call you shared, listed by decreasing probability:
import json
with open("kevintolan/savs2.1_proba.json") as file:
bvd_json = json.load(file)
bvd_proba = {
species["common_name"]: species["probability"]
for species in bvd_json["events"][0]["fine"].values()
}
print(sorted(bvd_proba.items(), key=lambda item: -item[1]))
So, "other" is the most probable species, followed by "American Tree Sparrow". The correct species (SAVS) only ranks as #6 in the list. Clearly, the species classifier is not reliable here.
However, not all hope is lost. We can query the taxonomical neural network at a different level of taxonomical hierarchy than the species level: in particular, BirdVox now integrates a family classifier.
with open("kevintolan/savs2.1_proba.json") as file:
bvd_json = json.load(file)
bvd_proba = {
species["common_name"]: species["probability"]
for species in bvd_json["events"][0]["medium"].values()
}
print(sorted(bvd_proba.items(), key=lambda item: -item[1]))
(note that i replaced "fine" by "medium" in the script above). Output:
[('American sparrow', 0.6484168767929077), ('other', 0.3515831232070923), ('Cardinal', 0.06819871068000793), ('Thrush', 0.05104416608810425), ('Warbler', 0.00031512975692749023)]
This time, the most probable result, "American sparrow", is
indeed the correct answer."
I hope this helps!
Sincerely,
Vincent.
