Issue with P(r) calculation in Raw v 2.1.1
5 views
Skip to first unread message
Wellington Claiton Leite
Jun 7, 2021, 12:42:49 PM6/7/21
to BioXTAS RAW
Dear,
I am having an issue when start the calculation of the P(r) in the RAW v 2.1.1. The error is posted bellow.
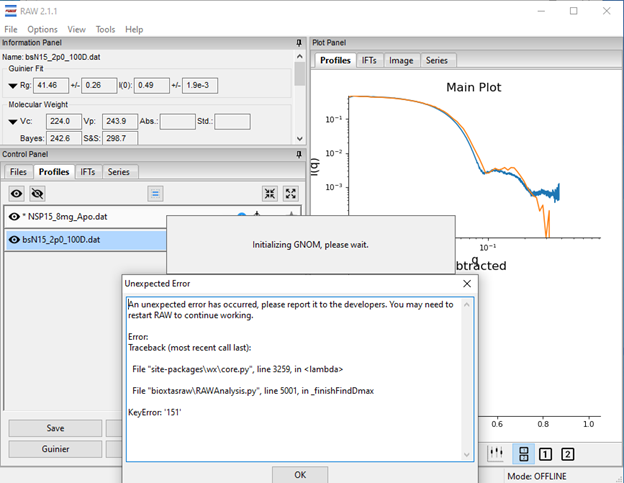
An unexpected error has occurred, please report it to the developers. You may need to restart RAW to continue working.
Error:
Traceback (most recent call last):
File "site-packages\wx\core.py", line 3259, in <lambda>
File "bioxtasraw\RAWAnalysis.py", line 4991, in _finishFindDmax
RuntimeError: wrapped C/C++ object of type GNOMControlPanel has been deleted
Jesse Hopkins
Jun 7, 2021, 12:58:25 PM6/7/21
to bioxt...@googlegroups.com
Hi Wellington,
That particular error would suggest that you're closing the IFT window before the initial automatic determination of Dmax is finished. Is that the case? If so, you should leave the window open until it displays a P(r) function.
If that's not the issue, please let me know what OS you're using, how you installed RAW (e.g. prebuilt or from source), and if possible send me the dataset that's causing this error so I can troubleshoot. A screenshot of RAW when you see the error message might also help.
All the best.
- Jesse
----
Jesse Hopkins, PhD
Beamline Scientist
BioCAT, Sector 18
Advanced Photon Source
--
You received this message because you are subscribed to the Google Groups "BioXTAS RAW" group.
To unsubscribe from this group and stop receiving emails from it, send an email to bioxtas_raw...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/21f9f3a1-e0e2-49b9-8e7b-f6068109c872n%40googlegroups.com.
Wellington Claiton Leite
Jun 18, 2021, 9:57:55 AM6/18/21
to bioxt...@googlegroups.com
Hi Jessie,
I apologize for the late response, I got busy with other analysis and could not be looking at this.
I have installed the latest version of RAW 2.1.1 in a Windows OS. Even when I wait until the initial automatic determination of Dmax gets done the same issue shows up. In the screenshot of the Raw windows there are two scattering profiles, the blue one is SAXS data and orange is SANS data, the error mostly happens when I have to analyze the SANS data. I have attached the scattering profiles.
Thank you for your help with this.
Best
Wellington

You received this message because you are subscribed to a topic in the Google Groups "BioXTAS RAW" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/bioxtas_raw/_otRubR_6t8/unsubscribe.
To unsubscribe from this group and all its topics, send an email to bioxtas_raw...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/CAGRN2W1ROdyFQWA%2BViqEZ%2B_g8aA1R2mGfnT_z5jP_NL9sM_26Q%40mail.gmail.com.
Wellington Claiton Leite
Jesse Hopkins
Jun 18, 2021, 10:19:00 AM6/18/21
to bioxt...@googlegroups.com
Hi Wellington,
It looks like you're using Windows? Is it Windows 10? I was unable to get that error message with either profile that you sent with RAW 2.1.1 on MacOS. If you let me know what your OS is, I'll test and see if it's platform specific.
Also, can you send me the config file you're using (if any)? It's possible it's something particular in your GNOM settings, so using identical conditions would be good.
All the best.
- Jesse
----
Jesse Hopkins, PhD
Deputy Director
BioCAT, Sector 18
Advanced Photon Source
To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/CAMyVvLqONUPWas3uo6MYgoJEPADvNdRvZTxoTUvGNSWH8xKLMw%40mail.gmail.com.
Wellington Claiton Leite
Jun 18, 2021, 10:32:20 AM6/18/21
to bioxt...@googlegroups.com
Yes, I am using Windows 10 Enterprise, and I am not using any specific configuration file, what I have is the same from the installer. I also screenshot my GNON configuration here. 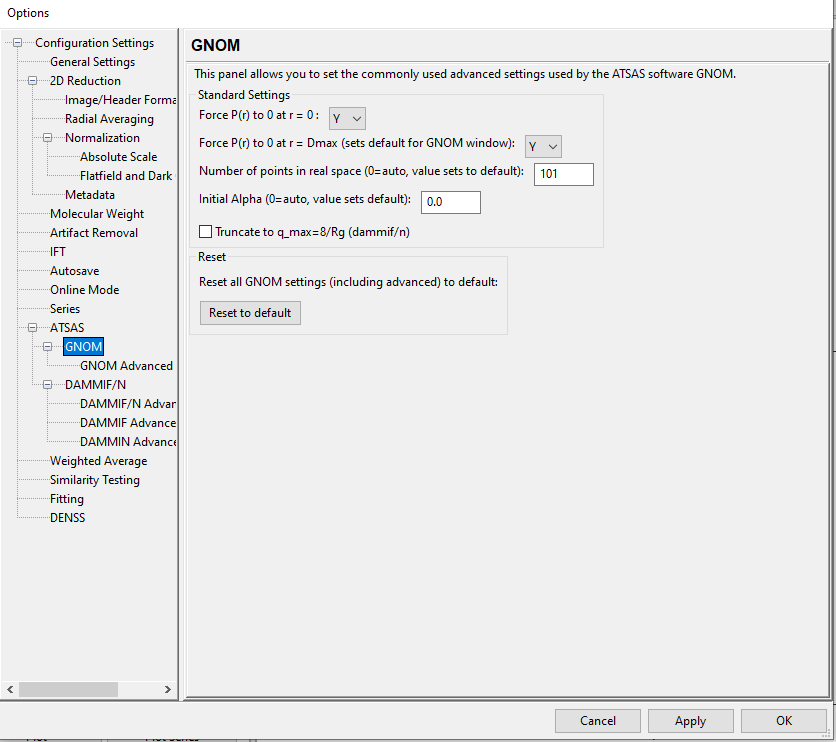


To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/CAGRN2W0geR7fthq__XaYJKRNF5TN3sUnAPvLuY5dx%3DBHxLPj_g%40mail.gmail.com.
Wellington Claiton Leite
Jesse Hopkins
Jun 18, 2021, 10:39:36 AM6/18/21
to bioxt...@googlegroups.com
Hi Wellington,
I see the problem in your config settings. In the first config screen you sent an image of, set 'Number of points in real space' to 0. RIght now it's set to more than the number of q points, which causes a GNOM error. It's likely that you at some point loaded a config from an older version of RAW that still used 101 as the default, instead of 0, and that's now being loaded by default when you open RAW.
To easily save the change when you next open RAW, just save the configuration once you make the change. Then RAW will automatically load that new config when it opens. Alternatively, when RAW asks to load the last used config file when it starts up, tell it 'No'.
Let me know if that fixes it.
All the best.
- Jesse
----
Jesse Hopkins, PhD
Deputy Director
BioCAT, Sector 18
Advanced Photon Source
To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/CAMyVvLo8gR17piAc%2BVNwBnEkKbui%2B1UySZwO5J57byC9QzTZwQ%40mail.gmail.com.
Wellington Claiton Leite
Jun 18, 2021, 12:24:20 PM6/18/21
to bioxt...@googlegroups.com
HI Jesse,
Yes, it works well. The issue was the configuration.
Thank you for your help with this.
Best
Wellington
To view this discussion on the web visit https://groups.google.com/d/msgid/bioxtas_raw/CAGRN2W0tBPTaD-JaR6pR-mQR8b5rp-PO-%3DX%3DRsyLxuExr0VSnA%40mail.gmail.com.
Wellington Claiton Leite
Reply all
Reply to author
Forward
0 new messages
