help
438 views
Skip to first unread message
Luiza Stoffel
Aug 23, 2022, 2:42:36 AM8/23/22
to Biogeme
hello!
has anyone had this problem? does anyone know how to help?
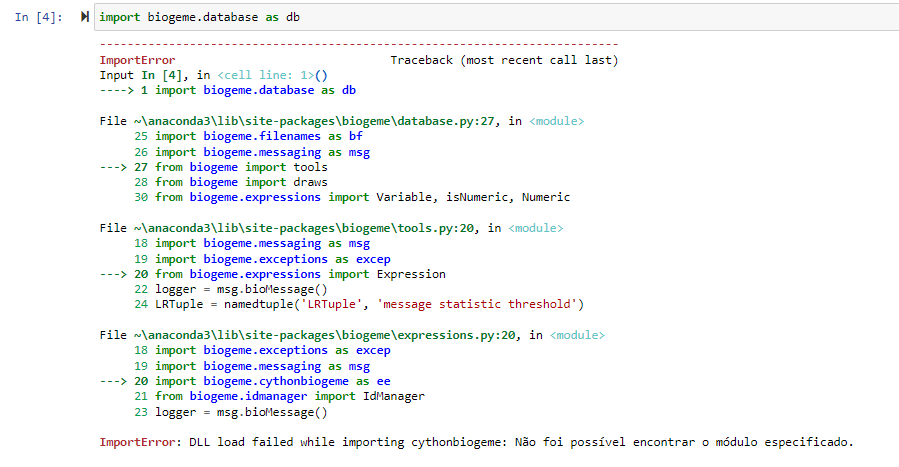
has anyone had this problem? does anyone know how to help?

thanks!
Bierlaire Michel
Aug 23, 2022, 3:01:41 AM8/23/22
to stoff...@gmail.com, Bierlaire Michel, Biogeme
On which platform is it? Windows?
On 23 Aug 2022, at 05:03, Luiza Stoffel <stoff...@gmail.com> wrote:
hello!
has anyone had this problem? does anyone know how to help?
<priny.png>
thanks!--
You received this message because you are subscribed to the Google Groups "Biogeme" group.
To unsubscribe from this group and stop receiving emails from it, send an email to biogeme+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/4218c142-867a-4b51-a187-0f1f68c673ben%40googlegroups.com.
<priny.png>
Bierlaire Michel
Aug 23, 2022, 10:14:43 AM8/23/22
to stoff...@gmail.com, Bierlaire Michel, Biogeme
Although they have been tested before being uploaded, it seems that the executables for Windows did not include a DLL.
I have now uploaded a new set of executables that include the DLL. I had to label them 3.2.10, although it is identical to 3.2.9.
It seems to have solved the problem.
Can you please confirm that it works for you?
Thanks,
Michel
On 23 Aug 2022, at 05:03, Luiza Stoffel <stoff...@gmail.com> wrote:
hello!
has anyone had this problem? does anyone know how to help?
Bierlaire Michel
Aug 23, 2022, 11:59:24 AM8/23/22
to Biogeme, Bierlaire Michel
Did you try these suggestions?
https://stackoverflow.com/questions/66060487/valueerror-numpy-ndarray-size-changed-may-indicate-binary-incompatibility-expOn 23 Aug 2022, at 16:45, stoff...@gmail.com wrote:
Now, this message appears:
"ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject"
<image.png>
Att,Luiza Stoffel
Rodrigo Tapia
Sep 12, 2022, 6:32:21 AM9/12/22
to Biogeme
Hi all,
Had the same issue (
"ValueError: numpy.ndarray
size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject"
) today and I did the following:
- pip uninstall biogeme
-
pip uninstall numpy
- pip install numpy
- conda update --all
- pip install biogeme
Steps 2 and 3 might be redundant.
This was done in a new laptop with Windows 11.
Hope this helps
Siti Fadilah
Sep 16, 2022, 3:34:22 AM9/16/22
to Biogeme
Dear all,
I just moved from macOS to Windows and the biogeme package has been installed very well. However, when I tried to run one of the examples attached to this URL:
Biogeme (epfl.ch), I also got the same error, even the NumPy itself worked!
I have tried countless times to uninstall and install the biogeme and NumPy, but it has not been solved yet.
Please help me to overcome this issue.
Thank you
Bierlaire Michel
Sep 20, 2022, 5:21:15 AM9/20/22
to srfad...@gmail.com, Bierlaire Michel, Biogeme
On 15 Sep 2022, at 10:30, Siti Fadilah <srfad...@gmail.com> wrote:
Dear all,
I just moved from macOS to Windows
That’s definitely not a good idea ;-)
and the biogeme package has been installed very well. However, when I tried to run one of the examples attached to this URL: Biogeme (epfl.ch), I also got the same error, even the NumPy itself worked!I have tried countless times to uninstall and install the biogeme and NumPy, but it has not been solved yet.
Create a brand new environment with python 3.9:
conda create -n myenv python=3.9 pip numpy
conda activate myenv
Then, install the fiollowing packages:
pip install pandas tqdm scipy cython biogeme
Note that python 3.10 should work as well, although some issues have been reported.
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/1d875669-a005-46bb-9d16-8ce6cac35b68n%40googlegroups.com.
<WhatsApp Image 2022-09-15 at 15.58.27.jpeg>
Siti Fadilah
Sep 21, 2022, 3:20:18 AM9/21/22
to Bierlaire Michel, Biogeme
Dear Prof. Bierlaire,
Finally, it works! Thank you for providing the solution.
Best regards,
Siti
Rim
Mar 7, 2023, 9:29:08 AM3/7/23
to Biogeme
Dear all,
Is anyone still encountering the same error message?
ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject
I was running the 01logit.py example code on windows with Python 3.9.12 and biogeme 3.2.10. I have already tested running the solutions mentioned above in the anaconda prompt but I still have the same error message.
Thank you in advance for any help!Rim
Thank you in advance for any help!Rim
Reply all
Reply to author
Forward
0 new messages
