Wikipathway Widget xref recognition
14 views
Skip to first unread message
jakob.v...@gmail.com
Jul 8, 2020, 2:56:23 PM7/8/20
to wikipathways-discuss
Dear team,
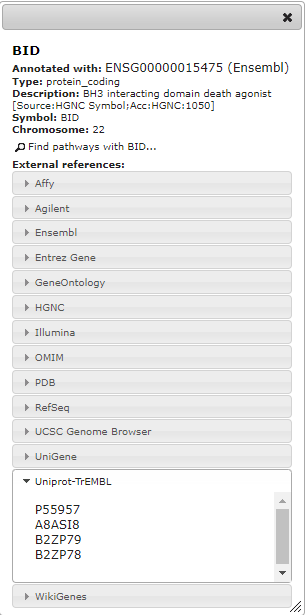
I have started using your excellent Widget for displaying protein fold change color-coded on the pathways. This in principle works great, but there is an issue with consistency of xrefs that somehow limits usability. As far as I understand colors are only displayed when the xref given in url matches to the value shown in "Associated with" field, not if it matches only to one of the other external references. In the attached image, only "xref= ENSG00000080546,Ensembl" would work, not "xref=Q9Y6P5,Uniprot-TrEMBL". Is this behavior intended?
As some nodes are associated with i.e. Ensembl and others with Uniprot-TrEMBL main xrefs this means that I have to supply both xrefs in the url which in turn can lead to very long urls and a Request-URI Too Long error form the server. My main identifiers are from Uniprot and I currently use BridgeDb for mapping between that and Entrez Gene as well as Ensembl which seems to cover all proteins.
Thanks a lot for your effort!
Cheers,
Jakob
Examples for this situation:
https://www.wikipathways.org/wpi/PathwayWidget.php?id=WP707&xref[]=P55957,Uniprot-TrEMBL&colors=green (doesn't work)
Egon Willighagen
Jul 8, 2020, 3:25:38 PM7/8/20
to wikipathways-discuss
Dear Jakob, all (developers, see my question at the end),
On Wed, Jul 8, 2020 at 8:56 PM <jakob.v...@gmail.com> wrote:
I have started using your excellent Widget for displaying protein fold change color-coded on the pathways. This in principle works great, but there is an issue with consistency of xrefs that somehow limits usability. As far as I understand colors are only displayed when the xref given in url matches to the value shown in "Associated with" field, not if it matches only to one of the other external references. In the attached image, only "xref= ENSG00000080546,Ensembl" would work, not "xref=Q9Y6P5,Uniprot-TrEMBL". Is this behavior intended?
The mapped identifiers on the website, as with the screenshot you posted, are dynamically looked up from the BridgeDb webservice. I do not think the widget has the functionality.
So, "intended" is likely a fair description: you have to use the identifier used in the GPML.
As some nodes are associated with i.e. Ensembl and others with Uniprot-TrEMBL main xrefs this means that I have to supply both xrefs in the url which in
turn can lead to very long urls and a Request-URI Too Long error form the server. My main identifiers are from Uniprot and I currently use BridgeDb for
mapping between that and Entrez Gene as well as Ensembl which seems to cover all proteins.
All, didn't we have some discussion at some point about GPML copies with normalized identifiers? For example, all gene identifiers in xrefs in the GPML all converted to Ensembl or so?
Egon
Hi, do you like citation networks? Already 51% of all citations are available available for innovative new uses. Join me in asking the American Chemical Society to join the Initiative for Open Citations too. SpringerNature, the RSC and many others already did.
-----
E.L. Willighagen
Department of Bioinformatics - BiGCaT
Maastricht University (http://www.bigcat.unimaas.nl/)
Homepage: http://egonw.github.com/
Blog: http://chem-bla-ics.blogspot.com/
PubList: https://www.zotero.org/egonw
ORCID: 0000-0001-7542-0286
ImpactStory: https://impactstory.org/u/egonwillighagen
Department of Bioinformatics - BiGCaT
Maastricht University (http://www.bigcat.unimaas.nl/)
Homepage: http://egonw.github.com/
Blog: http://chem-bla-ics.blogspot.com/
PubList: https://www.zotero.org/egonw
ORCID: 0000-0001-7542-0286
ImpactStory: https://impactstory.org/u/egonwillighagen
Jakob Vowinckel
Jul 10, 2020, 3:55:29 PM7/10/20
to wikipathways-discuss
Dear Egon,
thank you very much for your message! I am now using a method previously suggested in this group by first creating a mapping table from GraphIds to UniProt, Ensembl, and Gene Id, and then calling the widget with the GraphIds. This works reasonably well. It would still be nice to have a unified xref (at least within one pathway) to be able to avoid that step. Again, congrats to the great widget.
Cheers, Jakob
Anders Riutta
Jul 10, 2020, 4:24:48 PM7/10/20
to wikipathways-discuss
Hi Jakob,
We have another version of the viewer in development. It maps gene product identifiers to all of the following: Ensembl, Entrez (NCBI) Gene, HGNC and Wikidata. If you'd like to specify just Ensembl identifiers, you can do so like this (note the underscores):
https://pathway-viewer.toolforge.org/?id=WP707&green=Ensembl_ENSG00000015475,Ensembl_ENSG00000035928
Note that the pathway author specified Entrez (NCBI) Gene 5981 for RFC (on the right), but the highlighter accepted the Ensembl identifier: Ensembl_ENSG00000035928.
The green color in the development viewer is different from the one in the current viewer, but you can specify a lighter green as a hex code like this:
Even though it's in development, this viewer should be reasonably stable.
Best,
Anders
--
You received this message because you are subscribed to the Google Groups "wikipathways-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to wikipathways-dis...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/wikipathways-discuss/77def0d4-8ce8-4799-b5b6-e7aac1847806n%40googlegroups.com.
--
Anders Riutta
Gladstone Institutes | Science Overcoming Disease
Ondrej Kuda
Aug 24, 2020, 7:16:53 AM8/24/20
to wikipathways-discuss
Dear Egon,
I would like to use GPML pathways with the normalized identifiers. Is there a way how to convert all xrefs in the GPML file into HMDB for offline use? Or would it be possible to update the repository with normalized identifiers? The ecosystem of identifiers is so complex that some kind of unification would be great.
Thank you,
Ondrej
On Wednesday, July 8, 2020 at 9:25:38 PM UTC+2 egon.wil...@gmail.com wrote:
Anders Riutta
Aug 24, 2020, 12:26:40 PM8/24/20
to wikipathways-discuss
Hi Ondrej,
We have another version of the
viewer in development. It maps gene product identifiers to all of the
following: Ensembl, Entrez (NCBI) Gene, HGNC and Wikidata. For example, if you'd like
to specify just Ensembl identifiers, you can use link (note the
underscores):
https://pathway-viewer.toolforge.org/?id=WP707&green=Ensembl_ENSG00000015475,Ensembl_ENSG00000035928
Note
that the pathway author specified Entrez (NCBI) Gene 5981 for RFC (on
the right), but the highlighter accepted the Ensembl identifier:
Ensembl_ENSG00000035928.
The green color in the
development viewer is different from the one in the current viewer, but
you can specify a lighter green as a hex code like this:
Even though it's in development, this viewer should be reasonably stable.
Best,
Anders
--
You received this message because you are subscribed to the Google Groups "wikipathways-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to wikipathways-dis...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/wikipathways-discuss/a6df7303-3fa5-4402-b014-70d85628c76en%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
