Gene Symbol Aliases Problem
20 views
Skip to first unread message
pchev...@gmail.com
Jul 14, 2020, 8:09:28 PM7/14/20
to wikipathways-discuss
Hi,
Sorry if this is an issue that has been discussed before. There seems to be an issue when you search on a gene symbol (e.g. IL6 versus IL-6). Both gene symbols return two different sets of pathways but since they are they are aliases of each other they should return all of the same (a union of IL6 and IL-6) pathways. Could you create an alias file used in the search that would fix this issue?
Thanks,
Phil
search IL6 -> 115 pathways returned (Pathogenesis of SARS-CoV-2 Mediated by nsp9/nsp10 Complex (Homo sapiens) is missing_
search IL-6 -> 15 pathways returned (Pathogenesis of SARS-CoV-2 Mediated by nsp9/nsp10 Complex (Homo sapiens) is now present)
Martina Summer-Kutmon
Jul 15, 2020, 5:33:09 AM7/15/20
to wikipathways-discuss
Dear Phil,
I'll look into this issue and check if you can do an actual identifier search using the search field on the website (I think so but I am uncertain of the syntax right now).
Currently, you are using the free text search available on the website, which is looking for the text in the pathways but also descriptions, titles and literature references.
Using the following search returns the union of the two searches.
IL6 OR IL-6
If you use IL6 AND IL-6 you can see the 4 pathways that use both spelling versions.
If you want to look for pathways that contain the gene of interest as an actual datanode (element in the pathway), I would advise to use the webservice, PathVisio plugin (search by Xref) or R package (see example code below). You will see that there are 59 pathways containing 103 IL6 datanodes (e.g. WP4122 contains 17 IL-6 datanodes as participants in different complexes).
Hope this helps? Please let me know if you need further information.
Best regards,
Tina
library(dplyr)
# looking for pathways containing the human IL6 gene
res <- as.data.frame(rWikiPathways::findPathwayIdsByXref("ENSG00000136244","En"))
res <- as.data.frame(rWikiPathways::findPathwayIdsByXref("ENSG00000136244","En"))
colnames(res) <- "pathway"
# some pathways contain multiple IL6 data nodes - those will be grouped and counted
res.grouped <- res %>% group_by(pathway) %>% summarize(count = n())
--
You received this message because you are subscribed to the Google Groups "wikipathways-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to wikipathways-dis...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/wikipathways-discuss/b50d9abf-d950-4116-b30c-acf11d116a90o%40googlegroups.com.
Phil Chevalier
Jul 20, 2020, 7:18:18 PM7/20/20
to wikipathwa...@googlegroups.com
Dear Tina,
Thanks for your quick response. The current issue is that Wikipathways does not require the use of the official gene symbols (HGNC) for datanodes in pathways. This can result in a datanode using an alias instead of an official gene symbol which would result in cases where not all the expected pathways returned. This is a broad issue not just for gene symbols IL6 and IL8. There are many other examples: MYC/C-MYO, GAPD/GAPDH, PRKAA2/AMPK and etc. I understand that Wikipathways is an open ended community supported site and a controlled vocabulary (official gene symbols) is not strictly enforced but I believe this issue needs to be addressed. Possible solutions are:
1) Update Wikipathways to use the official gene symbols.
2) Update Wikipathways to use an alias file (like in the free text search - in addition to descriptions, titles and literature references all the pathways could be returned).
To fix my specific issue I am going through “Instructions for WikiPathways Authors” to learn how to either edit the gene symbol on the current pathway or copy the existing pathway and then add the official gene symbol.
Thank you,
Phil
You received this message because you are subscribed to a topic in the Google Groups "wikipathways-discuss" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/wikipathways-discuss/xvKd4hqIkuE/unsubscribe.
To unsubscribe from this group and all its topics, send an email to wikipathways-dis...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/wikipathways-discuss/CAA2qCtU7pGpdc%2BrY6qMB86js%3Db1S8cqbABOxZnzzmqT5QsyVSw%40mail.gmail.com.
Martina Summer-Kutmon
Jul 22, 2020, 4:43:30 AM7/22/20
to wikipathways-discuss
Dear Phil,
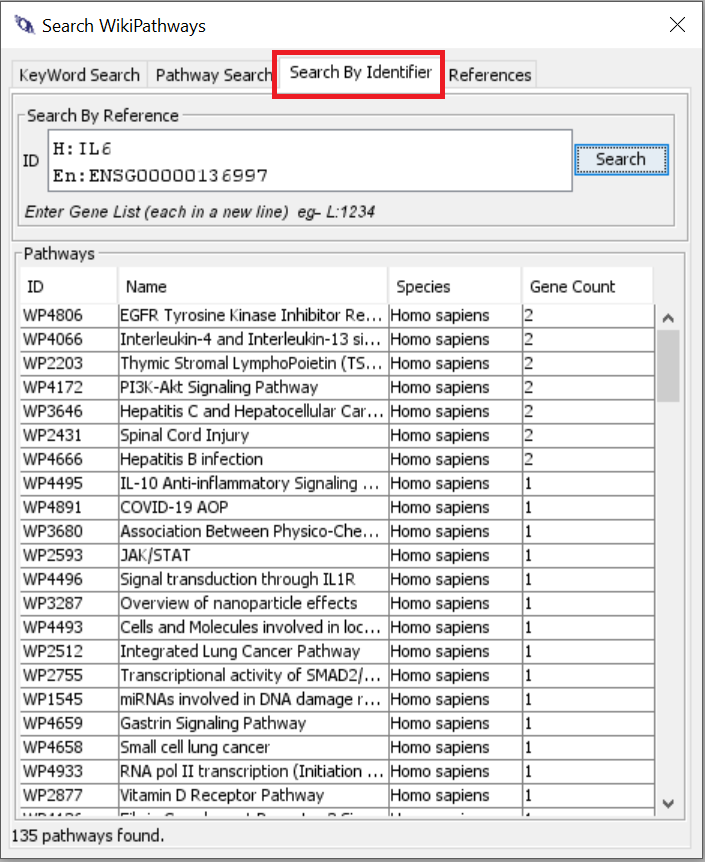
So in WikiPathways, every data node has a label and an identifier. For searches like you want, I would always advise to use the identifier search and not by text / label. For human genes, you can actually use the official gene symbols (HGNC) since we have an integrated identifier mapping service (BridgeDb).
Unfortunately, the search syntax for search by identifier does not work in the text field on the website. I will try to fix that in a future release. So for now, you would need to use the PathVisio plugin (slower than the others - see screenshot below), the webservice or the R package.
Unfortunately, the search syntax for search by identifier does not work in the text field on the website. I will try to fix that in a future release. So for now, you would need to use the PathVisio plugin (slower than the others - see screenshot below), the webservice or the R package.
Hope that helps!
Best, Tina
PathVisio - with WikiPathways plugin installed > Search > Search by Identifier (one identifier per line - as an example IL6 and MYC) - the top pathways in the result contain both of the provided genes.

To view this discussion on the web visit https://groups.google.com/d/msgid/wikipathways-discuss/3070E3DC-58D0-4AE5-8966-30362AE3A4A6%40gmail.com.
Reply all
Reply to author
Forward
0 new messages
