Is there a way to report the area occupied by each classification?
250 views
Skip to first unread message
Vetdocjim1
Feb 17, 2018, 4:17:18 PM2/17/18
to QuPath users
Colleagues:
Once the tissue has been classified, is there a way to report the area in um2 occupied by stroma (green below) and tumor (red below)?
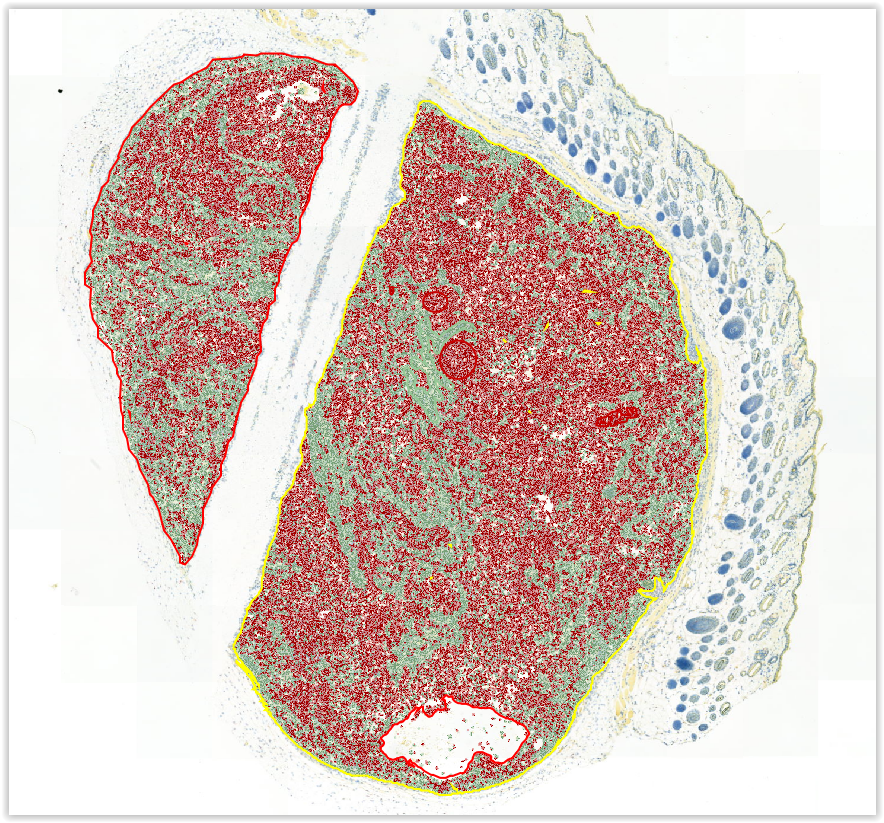
The program nicely reports the number of cells included in each classification and the total area measured, but it would be great to understand the area occupied by each classification.
Thanks for this program and for your help!
Jim
Once the tissue has been classified, is there a way to report the area in um2 occupied by stroma (green below) and tumor (red below)?
The program nicely reports the number of cells included in each classification and the total area measured, but it would be great to understand the area occupied by each classification.
Thanks for this program and for your help!
Jim
micros...@gmail.com
Feb 17, 2018, 5:32:54 PM2/17/18
to QuPath users
One problem with doing that in general is that it is somewhat dependent on the cell expansion selected when generating cells, so take some care in how you use the value.
That said, modified for your class (in my case, "Positive"), you can use a simple script to calculate what I think you are asking for:
Duplicate for as many classes as you are interested in.
That said, modified for your class (in my case, "Positive"), you can use a simple script to calculate what I think you are asking for:
def desiredCells = getDetectionObjects().findAll{it.getPathClass() == getPathClass("Positive")}
double totalArea = 0
for (def cell in desiredCells){
totalArea += cell.getMeasurementList().getMeasurementValue("Cell: Area")
}
println("Total Positive Area is: " + totalArea)
println("Mean area for Positive is: " + totalArea/desiredCells.size)
Duplicate for as many classes as you are interested in.
micros...@gmail.com
Feb 17, 2018, 5:52:44 PM2/17/18
to QuPath users
Expanding that out slightly for fun, the following adds the area value to each annotation in your image:
https://gist.github.com/Svidro/68dd668af64ad91b2f76022015dd8a45#file-cell-summary-measurements-to-annotation-groovy
https://gist.github.com/Svidro/68dd668af64ad91b2f76022015dd8a45#file-cell-summary-measurements-to-annotation-groovy
micros...@gmail.com
Feb 17, 2018, 5:57:04 PM2/17/18
to QuPath users
I should probably point out that the second script will only treat each cell as belonging to one annotation, so your annotations used for your classifier should be removed, or else they will get "credit" for the cells within them, not the parent annotation (I think). Alternatively, instead of cycling through all annotations, you could alter it to just work on the selected annotation, though I think it would likely still ignore any cells within sub-annotations.
Finn
Feb 19, 2018, 11:34:36 AM2/19/18
to QuPath users
Another solution I've found to this is to train tissue detection based on superpixels rather than cells, so that the tissue detection isn't contingent on a cell being detected or the cell expansion. The superpixels can be collapsed into annotations with 'Tile classifications to annotations' command, and will have a associated areas in the annotation measurements... Cells can then be detected inside the different annotation classes if needed.
micros...@gmail.com
Feb 19, 2018, 2:03:19 PM2/19/18
to QuPath users
I agree that that would be a more accurate way to handle measuring different tissue areas, provided the tissue is amenable to easy classification and superpixels! It nicely bypasses the problem of cell expansion and empty spaces between cells.
Pete
Feb 20, 2018, 8:34:55 AM2/20/18
to QuPath users
If you are feeling brave, you could try this script:
I've just written it for this purpose, although only tested it on a relatively small example (with ~100 000 cells). Nevertheless, it should generate annotations based upon already-detected cells, from which area measurements can be extracted - no superpixels necessary.
Be sure to check out the comments at the top, which give some extra information.
I think both approaches are valid; which one is better probably depends on whether you can get a more accurate superpixel classifier or a more accurate cell classifier.
lucia.m...@kcl.ac.uk
Oct 23, 2018, 11:15:20 AM10/23/18
to QuPath users
Hi Finn, I am trying to do something similar: detecting tumour areas and analyzing positive cells inside that, how do you classify superpixels?
Best,
Lucia
Finn
Oct 23, 2018, 12:08:25 PM10/23/18
to QuPath users
Hi Lucia --
You can create superpixels using the tiles and superpixels commands in the Analyze -> Region Identification pull down. There are a couple of algorithms (DOG/SLIC) so in the past I've played with the parameters of each until I get superpixels that delineate the boundaries I'm interested in... Superpixels are created without any associated measurements/features, so you'll need to add these for classification using the options in the Analyze -> calculate features menu. There are a lot of choices, so again you'll need some trial and error to see which are most helpful in your setting. Then you can train a classifier on the superpixels just like you would cells.
Once classified, you can collapse the superpixels to annotations, and do cell detection separately within annotation classes. I'm not sure Pete really intended this use, and this step (collapsing) can require a lot of computation.
In my experience, superpixels are noisier and a bit harder to classify than cells, but have worked really well on some projects. You might also have better luck with Pete's script (above) that does similar things based on cells...
Finn
micros...@gmail.com
Oct 23, 2018, 12:57:29 PM10/23/18
to QuPath users
Pete's final response here might be useful as well: https://groups.google.com/forum/#!searchin/qupath-users/Tile$20classifications$20to$20annotations%7Csort:date/qupath-users/gm0YYJxSriA/_iHs3ViaBgAJ
I have had the most luck using a script to manually classify the tiles, like a modification of : https://gist.github.com/Svidro/5b016e192a33c883c0bd20de18eb7764#file-a-simple-cell-classifier-groovy
There are a few other classifier scripts as well on that page, mostly taken from various places Pete has posted them. You would want to replace getCellObjects with getDetectionObjects in your case. One thing to be very careful about is the mu symbol and getting the exact string correct for your measurement label. This can be tricky due to exact spacing in some cases, and I would recommend this script if you have any trouble:
You can use a trained classifier as well, just like the example in the Wiki, but with the superpixels inside your classified annotations instead of cells: https://github.com/qupath/qupath/wiki/Classifying-objects
More information here: https://github.com/qupath/qupath/wiki/Object-classifications
Reply all
Reply to author
Forward
0 new messages
