Bio-Formats extension update - better support for more image formats in QuPath
1,310 views
Skip to first unread message
Pete
Jan 18, 2018, 1:47:25 PM1/18/18
to QuPath users
Hi everyone,
There has just been a new release of the QuPath Bio-Formats Extension - https://github.com/qupath/qupath-bioformats-extension
This is a fairly major rewrite, which aims to greatly improve the support for different image types in QuPath.
Previously, the fabulous Bio-Formats library was not being used to its full in QuPath, and hopefully this latest update will address this and makes many more images accessible.
Users of three file formats in particular should see significant improvements: CZI, NDPIS and VSI.
- Bio-Format’s memoization is now used. Accessing large files that were previously opened at least once is now much faster.
- QuPath tries to be smarter about how it uses image readers:
- QuPath will reuse large, memory-hungry file readers where it can, to reduce the risk of out-of-memory errors (consequently, some CZI that previously failed to open now work smoothly)
- QuPath will create multiple fast, lightweight readers for suitable images, and use them to request multiple image tiles in parallel to improve performance
- NDPIS files should open directly. Previously, only individual NDPI channels could be opened.
- A strict criterion checking the sizes of the different images within the image pyramid was relaxed; this makes it possible to open some VSI images that previously failed
- A bug related to resizing non-RGB images has been fixed; this could also prevent some other files from opening.
An important point about memoization is that it involves writing a (hidden) cache file whenever each image is opened. In some cases this file is extremely small, in others it could be as much as a gigabyte (and take a while to generate). By default, these files stored alongside the corresponding original images, in the same directories. Sometimes you might not want that (e.g. if reading files from a network share).
With that in mind, there are now specific Bio-Formats options available in the QuPath preferences.
Here you can turn memoization on or off, or specify another directory where the cache files should be written. You can also turn parallel file access off, if you have reason to believe it might be causing performance or memory trouble. Or if you just want to compare and see what it does.
The preferences also give you more control over when and where Bio-Formats is used. File extensions can be specified to make Bio-Formats the preferred reader or not. For example, if Bio-Formats is installed then it will be used to read NDPI files - but if you would prefer to use OpenSlide instead, then you can control that here.
Hopefully these updates are useful, but user feedback is really valuable to find out what works and what doesn't. This includes not only images that don't open, but also images that open strangely or may have errors in the metadata (e.g. the pixel size in microns).
For specific troublesome images, it is very helpful to first check 1) if the format is supported by Bio-Formats (i.e. on the list at https://docs.openmicroscopy.org/bio-formats/5.7.2/supported-formats.html), and 2) if it works using Bio-Formats in Fiji (www.fiji.sc).
If the answer to both of these is 'yes', then it's likely to be a QuPath-specific problem. Please report it here or at https://github.com/qupath/qupath-bioformats-extension/issues so it can be fixed.
Thanks!
Pete
David
Jan 19, 2018, 5:09:52 AM1/19/18
to QuPath users
Hi Pete,
thanks a lot for this update.
I just tried to find the new jar file and could not find it. Is the new extension another format?
The last qupath bioformat extension was a 15.9 (?) kb large .jar file that I drag and dropped onto the QuPath surface.
qupath-extension-bioformats.jar
If I follow the link
https://github.com/qupath/qupath-bioformats-extension
and then click the second here where we shall find the new jar file, I did not find it.
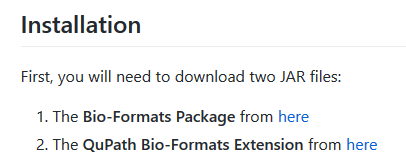
Ending up here, with the newest entry on 13th Oct 2016.
Did I maybe look at the wrong place for the jar file?
Best
David
thanks a lot for this update.
I just tried to find the new jar file and could not find it. Is the new extension another format?
The last qupath bioformat extension was a 15.9 (?) kb large .jar file that I drag and dropped onto the QuPath surface.
qupath-extension-bioformats.jar
If I follow the link
https://github.com/qupath/qupath-bioformats-extension
and then click the second here where we shall find the new jar file, I did not find it.
Ending up here, with the newest entry on 13th Oct 2016.
Did I maybe look at the wrong place for the jar file?
Best
David
Pete
Jan 19, 2018, 5:13:21 AM1/19/18
to QuPath users
Yes, but my fault for the link being out-of-date - sorry.
I've fixed it now. It should take you to https://github.com/qupath/qupath-bioformats-extension/releases
David
Jan 19, 2018, 5:13:57 AM1/19/18
to QuPath users
right now I found this link with the new jar file.
Maybe connect the other link to this place as well?
https://github.com/qupath/qupath-bioformats-extension/releases/tag/v0.0.4
Maybe connect the other link to this place as well?
https://github.com/qupath/qupath-bioformats-extension/releases/tag/v0.0.4
David
Jan 19, 2018, 5:39:15 AM1/19/18
to QuPath users
Thanks Pete.
I installed it by directly copying the .jar into the extensions folder. It works! :-)
The way before by drag and drop onto the QuPath User Interface did not work. Maybe because there was already a .jar file with the same name.
I got this message:
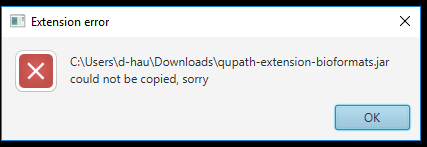
I installed it by directly copying the .jar into the extensions folder. It works! :-)
The way before by drag and drop onto the QuPath User Interface did not work. Maybe because there was already a .jar file with the same name.
I got this message:
Pete
Jan 19, 2018, 5:55:56 AM1/19/18
to QuPath users
Good!
I've also seen that message before on Windows. I guess it doesn't like overwriting the existing file while QuPath is running. The message didn't appear when I tried it on a Mac.
For anyone else having this trouble, the default extensions directory on Windows is usually under something like C:/Users/YourUsername/QuPath/extensions
You should be able to remove extensions / add new extensions by simply replacing the files in that directory. QuPath should discover them then whenever it is started up.
In case there is any trouble finding the correct directory, you can also request QuPath to open it for you:
import qupath.lib.gui.helpers.DisplayHelpers
import qupath.lib.gui.prefs.PathPrefs
import java.awt.Desktop
// Get the directory
def pathExtensions = PathPrefs.getExtensionsPath()
def dirExtensions = pathExtensions == null ? null : new File(pathExtensions)
print 'Extensions directory: ' + pathExtensions
// Check it is valid
if (dirExtensions == null || !dirExtensions.exists() || !dirExtensions.isDirectory()) {
DisplayHelpers.showErrorMessage("Open extensions directory", "The extensions directory could not be found!")
return
}
// Request the directory to be opened in Finder/Explorer
Desktop.getDesktop().open(dirExtensions)You'll might need to close QuPath after running this command so that you are able to change the files in the directory.
micros...@gmail.com
Jan 19, 2018, 2:51:48 PM1/19/18
to QuPath users
I can confirm that error happening on Windows systems, but dropping the file into the extensions folder everything worked cleanly.
In addition, for anyone else looking, another way to find the extensions folder location is in Preferences-> Extensions Directory
In addition, for anyone else looking, another way to find the extensions folder location is in Preferences-> Extensions Directory
David
Jan 29, 2018, 4:23:06 AM1/29/18
to QuPath users
Since the new bio-formats update, QuPath generates .bfmemo files for each image file. They are saved in the same folder like the image.
What is inside it and are they essential? I ask, because it is generating quite numerous files.
What is inside it and are they essential? I ask, because it is generating quite numerous files.
Pete
Jan 29, 2018, 4:43:32 AM1/29/18
to QuPath users
These are the cache files referred to in the original post (the bits about memoization).
I guess on Windows they aren't actually hidden.
They aren't essential and you can delete them; they are regenerated if they are needed.
There is some more information at https://docs.openmicroscopy.org/bio-formats/5.7.3/developers/matlab-dev.html#improving-reading-performance
You can turn them off in the QuPath preferences. But if you do turn them off then the performance is likely to suffer (probably more for some file formats than others).
Alternatively, you should be able to choose to put them into a separate directory (specified in the QuPath preferences). However I suspect - but have not tested - you might possibly experience some trouble with file paths that are too long for Windows to handle if you put them deep into the file system... so if you do specify a directory, watch out for that and try to choose a directory with a short path.
By default they are turned on, but maybe it would be better to turn them off by default... what do you think?
Message has been deleted
micros...@gmail.com
Apr 26, 2018, 11:04:24 AM4/26/18
to QuPath users
I noticed this: https://github.com/qupath/qupath-bioformats-extension/issues/2
Are you sure both you and your colleague have the same BioFormats version and settings? BioFormats was just updated recently (3 days ago), as was Pete's Bioformats plugin a few weeks ago I think.
It also looks like some of the ndpi files can do strange things to the metadata when the file size is large http://openslide.org/formats/hamamatsu/
This link also suggests that sometimes you will get files that can't be opened by other software: https://www.imnc.in2p3.fr/pagesperso/deroulers/software/ndpitools/
Are NDPIS files a sequence or Z stack or something else? I was not too clear on the difference between those and the .ndpi files. If you are allowed to host the image somewhere for Pete to look at that might help.
Are you sure both you and your colleague have the same BioFormats version and settings? BioFormats was just updated recently (3 days ago), as was Pete's Bioformats plugin a few weeks ago I think.
It also looks like some of the ndpi files can do strange things to the metadata when the file size is large http://openslide.org/formats/hamamatsu/
This link also suggests that sometimes you will get files that can't be opened by other software: https://www.imnc.in2p3.fr/pagesperso/deroulers/software/ndpitools/
Are NDPIS files a sequence or Z stack or something else? I was not too clear on the difference between those and the .ndpi files. If you are allowed to host the image somewhere for Pete to look at that might help.
Abhishek Rawat
Jul 11, 2018, 4:41:22 PM7/11/18
to QuPath users
Hello All!
I was wondering if anyone has had any issues importing .jp2 files?
I have a couple of ..jp2 files which QUpath refuses to import (with no details of the error).
The images work fine with ImageScope, so I know that's not the problem.
On FIJI also I am unable to import the same. Here I get the following error message for one of the images.
java.lang.NegativeArraySizeException
at loci.formats.MinMaxCalculator.openBytes(MinMaxCalculator.java:259)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:334)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:334)
at loci.plugins.util.ImageProcessorReader.openProcessors(ImageProcessorReader.java:186)
at loci.plugins.in.ImagePlusReader.readProcessors(ImagePlusReader.java:422)
at loci.plugins.in.ImagePlusReader.readPlanes(ImagePlusReader.java:387)
at loci.plugins.in.ImagePlusReader.readImage(ImagePlusReader.java:282)
at loci.plugins.in.ImagePlusReader.readImages(ImagePlusReader.java:243)
at loci.plugins.in.ImagePlusReader.readImages(ImagePlusReader.java:221)
at loci.plugins.in.ImagePlusReader.openImagePlus(ImagePlusReader.java:116)
at loci.plugins.in.Importer.readPixels(Importer.java:149)
at loci.plugins.in.Importer.run(Importer.java:86)
at loci.plugins.LociImporter.run(LociImporter.java:78)
at ij.IJ.runUserPlugIn(IJ.java:221)
at ij.IJ.runPlugIn(IJ.java:185)
at ij.IJ.runPlugIn(IJ.java:174)
at HandleExtraFileTypes.openImage(HandleExtraFileTypes.java:499)
at HandleExtraFileTypes.run(HandleExtraFileTypes.java:72)
at ij.IJ.runUserPlugIn(IJ.java:221)
at ij.IJ.runPlugIn(IJ.java:185)
at ij.IJ.runPlugIn(IJ.java:174)
at ij.io.Opener.openWithHandleExtraFileTypes(Opener.java:515)
at ij.io.Opener.openImage(Opener.java:369)
at ij.io.Opener.openImage(Opener.java:242)
at ij.io.Opener.open(Opener.java:109)
at ij.io.Opener.openAndAddToRecent(Opener.java:291)
at ij.plugin.DragAndDrop.openFile(DragAndDrop.java:192)
at ij.plugin.DragAndDrop.run(DragAndDrop.java:159)
at java.lang.Thread.run(Thread.java:745)
For the second image it's
java.lang.IllegalArgumentException: Dimensions (width=77456 height=61264) are too large
at java.awt.image.SampleModel.<init>(SampleModel.java:130)
at java.awt.image.ComponentSampleModel.<init>(ComponentSampleModel.java:146)
at java.awt.image.PixelInterleavedSampleModel.<init>(PixelInterleavedSampleModel.java:87)
at java.awt.image.PixelInterleavedSampleModel.createCompatibleSampleModel(PixelInterleavedSampleModel.java:144)
at com.sun.media.imageioimpl.plugins.jpeg2000.J2KReadState.readAsRaster(J2KReadState.java:429)
at com.sun.media.imageioimpl.plugins.jpeg2000.J2KImageReader.readRaster(J2KImageReader.java:556)
at ome.codecs.services.JAIIIOServiceImpl.readRaster(JAIIIOServiceImpl.java:177)
at ome.codecs.JPEG2000Codec.decompress(JPEG2000Codec.java:296)
at ome.codecs.JPEG2000Codec.decompress(JPEG2000Codec.java:267)
at loci.formats.codec.WrappedCodec.decompress(WrappedCodec.java:132)
at loci.formats.codec.JPEG2000Codec.decompress(JPEG2000Codec.java:63)
at loci.formats.in.JPEG2000Reader.openBytes(JPEG2000Reader.java:182)
at loci.formats.ChannelFiller.openBytes(ChannelFiller.java:156)
at loci.formats.ChannelFiller.openBytes(ChannelFiller.java:148)
at loci.formats.ChannelSeparator.openBytes(ChannelSeparator.java:198)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:348)
at loci.formats.DimensionSwapper.openBytes(DimensionSwapper.java:249)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:348)
at loci.formats.MinMaxCalculator.openBytes(MinMaxCalculator.java:269)
at loci.formats.MinMaxCalculator.openBytes(MinMaxCalculator.java:260)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:334)
at loci.formats.ReaderWrapper.openBytes(ReaderWrapper.java:334)
at loci.plugins.util.ImageProcessorReader.openProcessors(ImageProcessorReader.java:186)
at loci.plugins.in.ImagePlusReader.readProcessors(ImagePlusReader.java:422)
at loci.plugins.in.ImagePlusReader.readPlanes(ImagePlusReader.java:387)
at loci.plugins.in.ImagePlusReader.readImage(ImagePlusReader.java:282)
at loci.plugins.in.ImagePlusReader.readImages(ImagePlusReader.java:243)
at loci.plugins.in.ImagePlusReader.readImages(ImagePlusReader.java:221)
at loci.plugins.in.ImagePlusReader.openImagePlus(ImagePlusReader.java:116)
at loci.plugins.in.Importer.readPixels(Importer.java:149)
at loci.plugins.in.Importer.run(Importer.java:86)
at loci.plugins.LociImporter.run(LociImporter.java:78)
at ij.IJ.runUserPlugIn(IJ.java:221)
at ij.IJ.runPlugIn(IJ.java:185)
at ij.IJ.runPlugIn(IJ.java:174)
at HandleExtraFileTypes.openImage(HandleExtraFileTypes.java:499)
at HandleExtraFileTypes.run(HandleExtraFileTypes.java:72)
at ij.IJ.runUserPlugIn(IJ.java:221)
at ij.IJ.runPlugIn(IJ.java:185)
at ij.IJ.runPlugIn(IJ.java:174)
at ij.io.Opener.openWithHandleExtraFileTypes(Opener.java:515)
at ij.io.Opener.openImage(Opener.java:369)
at ij.io.Opener.openImage(Opener.java:242)
at ij.io.Opener.open(Opener.java:109)
at ij.io.Opener.openAndAddToRecent(Opener.java:291)
at ij.plugin.DragAndDrop.openFile(DragAndDrop.java:192)
at ij.plugin.DragAndDrop.run(DragAndDrop.java:159)
I understand the evidence points to this being a problem with Bio-Formats, but just cehcking if anyone has faced a similar situation?
cheers!
abhishek.
Pete
Jul 16, 2018, 4:10:43 AM7/16/18
to QuPath users
I'm afraid I don't have much JPEG2000 experience... I'd try the combination of Fiji + Bio-Formats (as you have done), and also OpenSlide + Python to see if you've any luck with either of them.
If not, then I'd suggest following up with either the Bio-Formats or OpenSlide developers. I'm not entirely sure if OpenSlide is expected to handle JPEG2000 in its current form (since this issue is open, perhaps not), but the format is listed as supported for Bio-Formats here - so I'd try Bio-Formats first.
If you do manage to confirm that either library can handle the images, but they still fail in QuPath, then I can explore where the trouble is on the QuPath side.
Abhishek Rawat
Jul 16, 2018, 2:17:03 PM7/16/18
to QuPath users
Thanks Pete.
Ya, I figured this was not a QUpath problem.
It seems this is known bug in BioFormats which only affects .jp2 images with N_pixels > 2^31 - 1.
The bug seems to be open since the last 6 years so I have little hope they will be of any help.
I can open the image in ImageScope so I will just figure out from there.
cheers!
abhi.
Pete
Jul 17, 2018, 2:29:44 AM7/17/18
to QuPath users
Ah, that's unfortunate...
I see it arose again 12 months ago. I guess you could add your voice to those asking for this to be addressed; there's a forum and a mailing list. I went to the OME meeting this year for the first time and met some of the Bio-Formats team - they are very helpful! But I guess it's not a straightforward issue to solve.
Can you say where the original image came from, and is there any other export option? I'm curious as to whether this is likely to arise more often in the future...
Abhishek Rawat
Jul 17, 2018, 10:16:58 AM7/17/18
to QuPath users
Hey Pete,
The images came from a BioImagene Inc whole slide scanner to which I do not have access.
So not really possible to export them differently from the scanner itself. I will just have to use some other s/w to read in the .jp2 and dump out some other format that I can work with.
To answer you question about likelihood of such issues happening more frequently in the future: Since this issue impacts images > 2^31 -1 pixels and whole slide images often exceed this threshold, I would say that it is likely this will keep happening in the future.
That said JPEG2000 is not a very widely used standard so I guess most people will get by.....and that's why Bio-Formats team has not bothered with fixing this bug in the last 6 years :)
cheers!
abhi.
Mariam Gachechiladze
Mar 2, 2019, 12:25:23 PM3/2/19
to QuPath users
Dear Pete,
I have a problem opening whole slide images in VSI format in QuPath. I have installed bioformats extension but it still does not help.
Would you have any solution for that? Or may be you could think of potential problems and how can I overcome them.
Thank you and best wishes,
Mariam
Pete
Mar 2, 2019, 12:29:35 PM3/2/19
to QuPath users
I would need to know much more precisely what your problem is, and perhaps would also require some example images. I do not work with VSI images myself.
(All my attention currently is given to updating the entire software and I don't plan to update the Bio-Formats extension specifically any time soon. There are enough changes that it might be the case in the next release the problem goes away anyway, but I cannot be sure.)
Pete
Mar 2, 2019, 2:19:19 PM3/2/19
to QuPath users
I can confirm that the image opens fine in the upcoming milestone release, but should also work ok in v0.1.2 with the Bio-Formats extension.
In v0.1.2 QuPath opens the first image that it finds in the file, which happens to be a low resolution overview in this case. But if you add the image to a project (which is usually a better idea anyway, instead of working with individual files) then all images should be added and you should find one called 20x that should have higher resolution data.
I should mention that .vsi files are a bit awkward since they tend to give all images the same names inside (e.g. 20x), which isn't so good in a QuPath project, where names should be unique. However I've tried to improve this by generating an image names that consists of both the file name and the 'internal' name. You can see https://petebankhead.github.io/qupath/2018/03/19/qupath-updates.html for some improvements in that direction that will also be in the next release, but v0.1.2 doesn't do this.
Finally, if you want to access the image without creating a project, then click on the 'Image' tab on the left and you should see 'Image list' at the bottom. If you select that, then you get access to the other images stored in the file... including the higher-resolution 20x image.
Reply all
Reply to author
Forward
0 new messages

