WLS.V error with estimator WLSMV
848 views
Skip to first unread message
rico...@gmail.com
Feb 19, 2018, 5:18:37 AM2/19/18
to lavaan
Dear Professor Yves,
Thank you for a great package which has given me a lot of joy! I have modeled some ordered data and would like to reproduce the results without using the full data, but when I try I get this error:
Thank you for a great package which has given me a lot of joy! I have modeled some ordered data and would like to reproduce the results without using the full data, but when I try I get this error:
Error in Gamma[[g]] %*% WD : requires numeric/complex matrix/vector arguments
(There is also an error with DWLS:
Error in crossprod(Delta[[g]], lavsamp...@WLS.VD[[g]] * diff) : non-conformable arguments In addition: Warning messages:
1: In diff * diff * WLS.VD : longer object length is not a multiple of shorter object length 2: In lavsamp...@WLS.VD[[g]] * diff : longer object length is not a multiple of shorter object length
)
Could you give me some hints of what I am doing wrong? The code is provided below:
mod <- '
LV =~ i1+i2+i3+i4+i5
'
fit <- cfa(mod, data, ordered=names(data),
estimator="WLSMV",
parameterization='theta',
std.lv=T)
fitcov <- cfa(mod, ordered=names(data),
sample.cov=lavCor(data,ordered=names(data)),
sample.nobs=20000,
WLS.V=lavInspect(fit, what="wls.v"),
std.lv=T,
estimator="WLSMV")
Sincerely,
Rico
Yves Rosseel
Feb 22, 2018, 2:36:03 AM2/22/18
to rico...@gmail.com, 'Anna' via lavaan
On 02/19/2018 11:18 AM, rico...@gmail.com wrote:
> fitcov <- cfa(mod, ordered=names(data),
> sample.cov=lavCor(data,ordered=names(data)),
> sample.nobs=20000,
> WLS.V=lavInspect(fit, what="wls.v"),
> std.lv=T,
> estimator="WLSMV")
This will not work (for now) for two reasons:
> fitcov <- cfa(mod, ordered=names(data),
> sample.cov=lavCor(data,ordered=names(data)),
> sample.nobs=20000,
> WLS.V=lavInspect(fit, what="wls.v"),
> std.lv=T,
> estimator="WLSMV")
- as soon as you provide sample.cov, lavaan assumes continuous data
- you are missing an important ingredient: Gamma (which you can get from
lavInspect(fit, "gamma") (see the NACOV= argument).
Yves.
rico...@gmail.com
Feb 22, 2018, 5:03:03 AM2/22/18
to lavaan
Thank you! So if I understand you correctly, the code below should reproduce the analysis in a future version of lavaan when sample.cov accepts categorical data? This reproduction will require, the correlation matrix, weight matrix and gamma matrix?
fitcov <- cfa(model=mod,
sample.cov=lavCor(data,ordered=names(data)),
sample.nobs=20000,
parameterization='theta',
WLS.V=lavInspect(fit, "wls.v"),
NACOV=lavInspect(fit, "gamma"),
std.lv=T,
estimator="WLSMV")
Best,
Rico
fitcov <- cfa(model=mod,
sample.cov=lavCor(data,ordered=names(data)),
sample.nobs=20000,
parameterization='theta',
WLS.V=lavInspect(fit, "wls.v"),
NACOV=lavInspect(fit, "gamma"),
std.lv=T,
estimator="WLSMV")
Best,
Rico
Yves Rosseel
Feb 26, 2018, 7:02:15 AM2/26/18
to lav...@googlegroups.com
Yes.
On 02/22/2018 11:03 AM, rico...@gmail.com wrote:
> Thank you! So if I understand you correctly, the code below should
> reproduce the analysis in a future version of lavaan when sample.cov
> accepts categorical data? This reproduction will require, the
> correlation matrix, weight matrix and gamma matrix?
>
> fitcov <- cfa(model=mod,
> sample.cov=lavCor(data,ordered=names(data)),
> sample.nobs=20000,
> parameterization='theta',
> WLS.V=lavInspect(fit, "wls.v"),
> NACOV=lavInspect(fit, "gamma"),
> std.lv=T,
> estimator="WLSMV")
>
> Best,
> Rico
>
>
>
> Den torsdag 22 februari 2018 kl. 08:36:03 UTC+1 skrev Yves Rosseel:
>
> You received this message because you are subscribed to the Google
> Groups "lavaan" group.
> To unsubscribe from this group and stop receiving emails from it, send
> an email to lavaan+un...@googlegroups.com
> <mailto:lavaan+un...@googlegroups.com>.
> To post to this group, send email to lav...@googlegroups.com
> <mailto:lav...@googlegroups.com>.
> Visit this group at https://groups.google.com/group/lavaan.
> For more options, visit https://groups.google.com/d/optout.
On 02/22/2018 11:03 AM, rico...@gmail.com wrote:
> Thank you! So if I understand you correctly, the code below should
> reproduce the analysis in a future version of lavaan when sample.cov
> accepts categorical data? This reproduction will require, the
> correlation matrix, weight matrix and gamma matrix?
>
> fitcov <- cfa(model=mod,
> sample.cov=lavCor(data,ordered=names(data)),
> sample.nobs=20000,
> parameterization='theta',
> WLS.V=lavInspect(fit, "wls.v"),
> NACOV=lavInspect(fit, "gamma"),
> std.lv=T,
> estimator="WLSMV")
>
> Best,
> Rico
>
>
>
> Den torsdag 22 februari 2018 kl. 08:36:03 UTC+1 skrev Yves Rosseel:
>
> On 02/19/2018 11:18 AM, rico...@gmail.com <javascript:> wrote:
> > fitcov <- cfa(mod, ordered=names(data),
> > sample.cov=lavCor(data,ordered=names(data)),
> > sample.nobs=20000,
> > WLS.V=lavInspect(fit, what="wls.v"),
> > std.lv <http://std.lv>=T,
> > fitcov <- cfa(mod, ordered=names(data),
> > sample.cov=lavCor(data,ordered=names(data)),
> > sample.nobs=20000,
> > WLS.V=lavInspect(fit, what="wls.v"),
> > estimator="WLSMV")
>
>
> This will not work (for now) for two reasons:
>
> - as soon as you provide sample.cov, lavaan assumes continuous data
> - you are missing an important ingredient: Gamma (which you can get
> from
> lavInspect(fit, "gamma") (see the NACOV= argument).
>
> Yves.
>
> --
>
>
> This will not work (for now) for two reasons:
>
> - as soon as you provide sample.cov, lavaan assumes continuous data
> - you are missing an important ingredient: Gamma (which you can get
> from
> lavInspect(fit, "gamma") (see the NACOV= argument).
>
> Yves.
>
> You received this message because you are subscribed to the Google
> Groups "lavaan" group.
> To unsubscribe from this group and stop receiving emails from it, send
> an email to lavaan+un...@googlegroups.com
> <mailto:lavaan+un...@googlegroups.com>.
> To post to this group, send email to lav...@googlegroups.com
> <mailto:lav...@googlegroups.com>.
> Visit this group at https://groups.google.com/group/lavaan.
> For more options, visit https://groups.google.com/d/optout.
alinage...@gmail.com
Nov 2, 2018, 6:26:08 AM11/2/18
to lavaan
Dear Professor Yves
I have a question on this exact solution, as this might be my solution.
I do a cfa on ordinal data and polychoric correlations (called "cormat_efa_r" which is conducted beforehand with hetcor() of the polycor package).
My fit looks like this:
fit_efa_R <- cfa(model=efa_mr, sample.cov=cormat_efa_r , sample.nobs=nobs_r,
estimator ="WLSMV",
orthogonal=F, std.lv=F)
Which is requiring a "user provided WLS.V matrix"
I have seen here is ued WLS.V=lavInspect(fit, what="wls.v") to solve this problem. But what exactely is "fit" in my case?
I would highly appreciate your help.
Best,
Alina
Terrence Jorgensen
Nov 2, 2018, 9:42:30 AM11/2/18
to lavaan
I have seen here is ued WLS.V=lavInspect(fit, what="wls.v") to solve this problem. But what exactely is "fit" in my case?
I think that would only work if you used lavaan to fit the model to your raw data, as shown in the original post. But if you use lavCor() to estimate cormat_efa_r, you might be able to get the WLS.V matrix you need from that output. (I'd wait for Yves to reply, though.)
Terrence D. Jorgensen
Assistant Professor, Methods and Statistics
Research Institute for Child Development and Education, the University of Amsterdam
Alina Gerlach
Nov 2, 2018, 9:53:35 AM11/2/18
to lav...@googlegroups.com
Thank you for your fast reply, Mr. Jorgensen.
I do need to conduct the correlation matrices with hetcor(), as this is a method to detect heterogeneous correlation matrices. I have ordered variables but different kinds.
Is there a way to conduct the needed matrix manually?
And, yes, I'd wait for Yves reply too!
--
You received this message because you are subscribed to a topic in the Google Groups "lavaan" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/lavaan/KZFGMlCuCDo/unsubscribe.
To unsubscribe from this group and all its topics, send an email to lavaan+un...@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Terrence Jorgensen
Nov 8, 2018, 4:26:40 AM11/8/18
to lavaan
I do need to conduct the correlation matrices with hetcor(), as this is a method to detect heterogeneous correlation matrices. I have ordered variables but different kinds.
That is what lavCor() offers, too. Read the ?lavCor help page and run the examples to see how it works. Unlike hetcor(), which only returns SEs (square-roots of sampling variances), lavCor() can provide a lavaan object from which you can obtain (with the vcov() function) the full sample covariance matrix needed to calculate the robust test statistic (although only the diagonal is used for finding point estimates).
Terrence Jorgensen
Apr 4, 2019, 10:40:27 AM4/4/19
to lavaan
fitcov <- cfa(model=mod,
sample.cov=lavCor(data,ordered=names(data)),
sample.nobs=20000,
parameterization='theta',
WLS.V=lavInspect(fit, "wls.v"),
NACOV=lavInspect(fit, "gamma"),
std.lv=T,
estimator="WLSMV")
I'm flirting with two-stage estimation, and I wanted to post here for posterity that lavInspect(fit, "wls.v") is the inverse of lavInspect(fit, "gamma") when using full weighted least squares estimation (estimator = "WLS"). When using estimator = "DWLS" (lavaan's default for categorical data), lavInspect(fit, "wls.v") is the inverse of a version of lavInspect(fit, "gamma") with all off-diagonal elements set to zero.
# when estimator = "WLS"
lavInspect(fit, "wls.v") == solve(lavInspect(fit, "gamma"))
# when estimator = "DWLS", for less computationally demanding estimation
lavInspect(fit, "wls.v") == solve(diag(diag(lavInspect(fit, "gamma"))))
This might come in handy for anyone (like Alina) who has to obtain their N-times-the-ACOV matrix (NACOV) and weight matrix (WLS.V) from external software.
Peter Halpin
Nov 30, 2020, 5:42:41 PM11/30/20
to lavaan
Hello! I realize this thread is a bit old but have run into a similar situation and am interested to see if anyone has follow-ups to report or any feedback on the solution proposed below.
sample.nobs=20000,
parameterization='theta',
WLS.V=lavInspect(fit, "wls.v"),
NACOV=lavInspect(fit, "gamma"),
std.lv=T,
estimator="WLSMV")
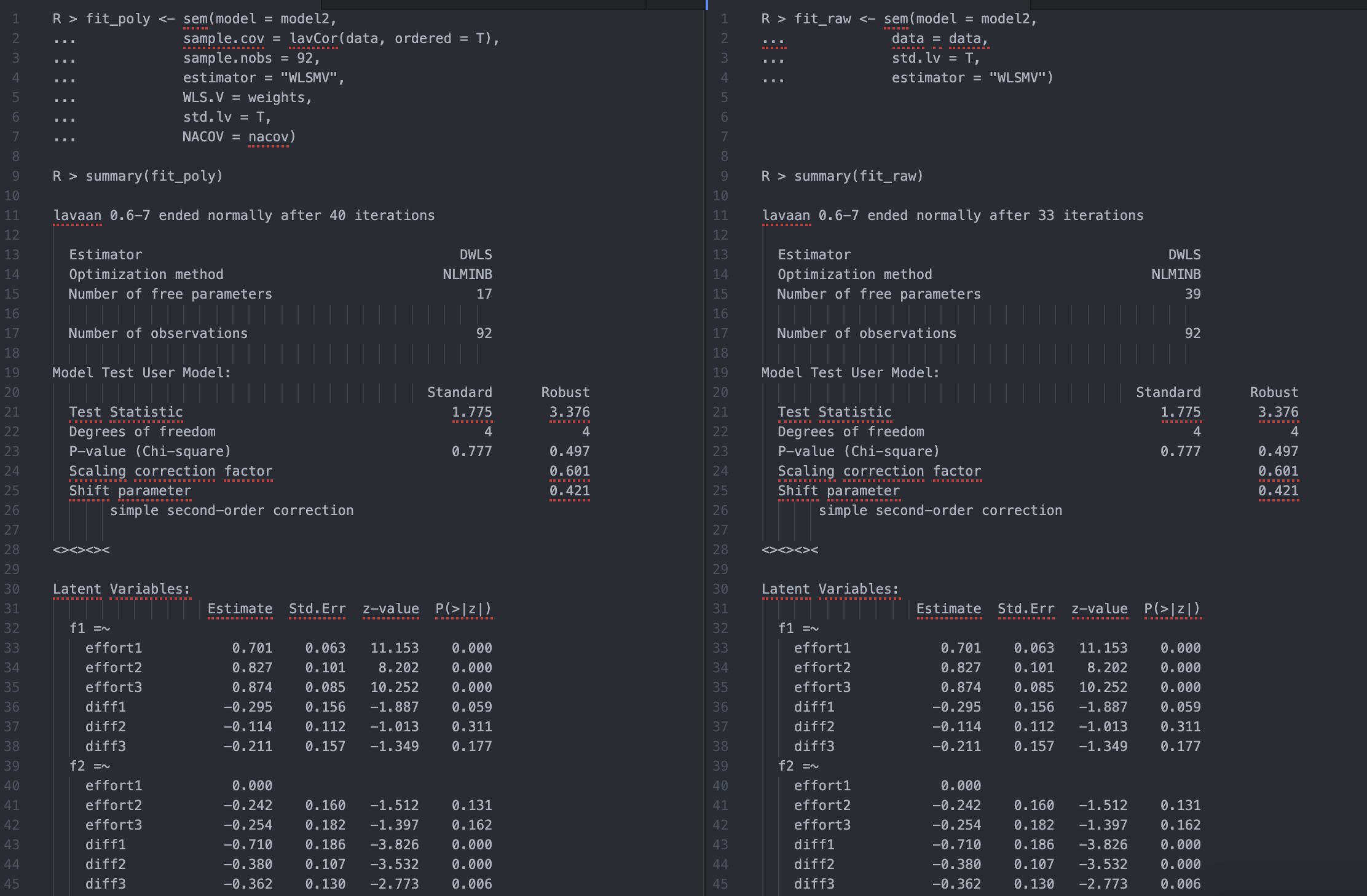
I am in a situation where I want to fit many models (> 200) to a large (80 X 80) polychoric correlation matrix. To reduce computational time, I would like to compute the polychorics only once and input them into lavaan.
This led me to essentially the same code posted above by Rico and Terrence above (reproduced here for clarity):
fitcov <- cfa(model=mod,
sample.cov=lavCor(data, ordered=names(data)),
sample.cov=lavCor(data, ordered=names(data)),
sample.nobs=20000,
parameterization='theta',
WLS.V=lavInspect(fit, "wls.v"),
NACOV=lavInspect(fit, "gamma"),
std.lv=T,
estimator="WLSMV")
where fit is a model run on the categorical raw data.
As Yves noted previously, this won't work because when sample.cov is used, lavaan assumes the data are continuous. I guess no update on that yet?
Anyway, when the data are continuous, this means that WLS.V and NACOV should be ordered as follows (taken from the help documentation for lavaan):
"The elements of the weight matrix should be in the following order (if all data is continuous): first the means (if a meanstructure is involved), then the lower triangular elements of the covariance matrix including the diagonal, ordered column by column." (quote_1)
On the other hand, the "wls.v" and "gamma" output from the categorical raw data fit will be organized for the categorical case:
"First the thresholds (including the means for continuous variables), then the slopes (if any), the variances of continuous variables (if any), and finally the lower triangular elements of the correlation/covariance matrix excluding the diagonal, ordered column by column." (quote_2)
The error that lavaan throws when running the cfa above is that the dimensions of WLS.V (and NACOV) are not conformable with the sample.cov. This is because they are obtained from the categorical raw data and therefore include the thresholds but do not include the diagonal elements of the covariance (polychoric correlation) matrix.
I am stubbornly still trying to make this work and I think I have found a solution. But I am not sure why (or if) it works in theory and am looking for any feedback. The solution is as follows.
We can get the elements of "wls.v" and "gamma" for the off-diagonal elements of the polychoric matrix using lavInspect with the raw data fit, as above. We can also order these as required in quote_1.
However, I am not sure what we could use for the elements of "wls.v" and "gamma", in place of the variances from quote_1? If anyone knows of relevant theory here, please send a reference!
Anyway, on a hunch I just used an identity (sub) matrix for the elements of "wls.v" and "gamma" for the variances. The output below compares the fit of this approach with the categorical raw data using a toy example (a 2-factor cfa with 6 ordinal indicators). The proposed approach gave the same GOF and results for the factor loadings, and even reproduced the point estimates for the residual variances (not shown). More surprising, the results of fit_poly did not change when I rescaled elements of "wls.v" and "gamma" for the variances by any constant.
I am tentatively making the following conclusions:
1. You can pass the polychorics to lavaan so long as the WLS.V and NACOV are organized as described in quote_1.
2. It doesn't matter what you use for the variances in quote_1, but an identity (sub) matrix works fine for them both. I have no theory to back this up.
Any input would be welcome! I would like to know if I am making a mistake before I try this with 200+ models!
Thanks, Peter

Terrence Jorgensen
Dec 4, 2020, 6:12:23 AM12/4/20
to lavaan
You can get everything you need from a fitted lavaan object, including the saturated model automatically fitted by lavCor()
fit_poly <- lavCor(data = Data, ordered = TRUE, output = "lavaan")
## obtain summary stats
sample.cov <- lavInspect(fit_poly, "sampstat")$cov
sample.mean <- lavInspect(fit_poly, "sampstat")$mean
sample.th <- lavInspect(fit_poly, "sampstat")$th
attr(sample.th, "th.idx") <- lavInspect(fit_poly, "th.idx")
sample.nobs <- lavInspect(fit_poly, "nobs")
WLS.V <- lavInspect(fit_poly, "WLS.V")
NACOV <- lavInspect(fit_poly, "gamma")
## fit model to summary stats
fit <- cfa(model, sample.cov = sample.cov, sample.mean = sample.mean,
WLS.V = WLS.V, NACOV = NACOV)
Reply all
Reply to author
Forward
0 new messages
