Pfam domains raw data input fruitfly
Daniel Gonzalez
Gerardo Perez
Hello, Daniel.
Thank you for your interest in the Genome Browser and your question about raw data of Pfam domain identification.
We have a file that has the mapping of domains to the dm6 RefSeq proteins:
https://hgwdev-gperez2.gi.ucsc.edu/~gperez2/mlq/mlq_30019/data/dm6/bed/pfam/ucscPfam.tab
If you were looking for something different, then we would encourage you to reach out to the PFAM helpdesk (pfam...@ebi.ac.uk) and CC us (genom...@soe.ucsc.edu). We annotate the genome, not the proteins.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible Google Groups forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
Gerardo Perez
UCSC Genomics Institute
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CADd%2Be27uKmumsqb-TW6CeoMyvz3sgJrq_BLEwpXLEv%2BO99%3DhzA%40mail.gmail.com.
Daniel Gonzalez
Luis Nassar
Thank you for sharing those examples.
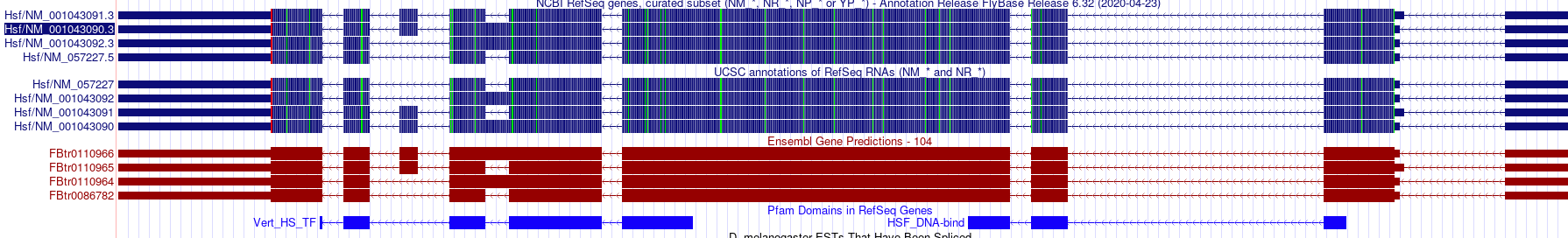
The first example, NM_166413.3, looks to be due to a bug in how we generate the alignments for this track. We have regenerated the track and it should now contain the two expected domains.
As far as the second example, that is an expected result due to how the track is generated. The pipeline calculates the domains for all the transcripts and then creates a final unique list which is the Pfam domains track. As you say, NP_001036555 only has a single domain, but the isoform NM_057227.5/NP_476575.1 results in the two domains seen on the pfam track can be verified on InterproScan.
I hope this is helpful. Please include gen...@soe.ucsc.edu in any replies to ensure visibility by the team. All messages sent to that address are archived on our public forum. If your question includes sensitive information, you may send it instead to genom...@soe.ucsc.edu.
Lou Nassar
UCSC Genomics Institute
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CADd%2Be26KnC-7Sd6CSNYzz1tGmqJo%3DOtmWRyF2xoxeyo82ehVBw%40mail.gmail.com.
