Re: [genome-www] Missing Annotation File
0 views
Skip to first unread message
Jairo Navarro Gonzalez
Sep 7, 2022, 8:34:12 PM9/7/22
to Charles-Alexandre Roy, gen...@soe.ucsc.edu, genom...@soe.ucsc.edu
Hello,
Thank you for using the UCSC Genome Browser and sending your inquiry.
The underlying data for the NCBI RefSeq Other track is stored inside of a bigBed file and not as a MySQL table. The table dump is a single line that points to the location of the bigBed file, which is the/gbdb file server (https://hgdownload.soe.ucsc.edu/downloads.html#gbdb).
You can download the bigBed file using the following URL:
http://hgdownload.soe.ucsc.edu/gbdb/hg38/ncbiRefSeq/ncbiRefSeqOther.bb
If you are not familiar with the bigBed file format, you can learn more about how to extract the data using the following help page:
I hope this is helpful. If you have any further questions, please reply to genom...@soe.ucsc.edu.
Jairo Navarro
UCSC Genome Browser
On Wed, Sep 7, 2022 at 4:29 PM Charles-Alexandre Roy <roy.charle...@gmail.com> wrote:
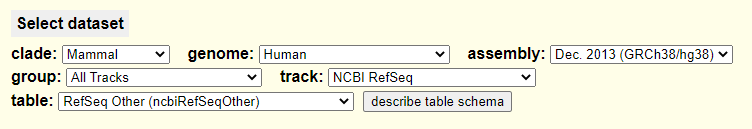
--Hello,I am writing because I've previously downloaded the RefSeq Other (ncbiRefSeqOther) file from the UCSC Table Browser with the following query:Going forward I'd like to automatically download the file from the goldenPath (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/database/ncbiRefSeqOther.txt.gz), but when I do the file only contains this line: /gbdb/hg38/ncbiRefSeq/ncbiRefSeqOther.bb. By contrast, the ncbiRefSeq.txt.gz file seems to be identical to the one I can obtain via the Table Browser. Is it possible that there's something wrong with how the ncbiRefSeqOther.txt.gz file is being dumped from the annotation database?Thank you for your time and help!Best,Charles
To unsubscribe from this group and stop receiving emails from it, send an email to genome-www+...@soe.ucsc.edu.
Reply all
Reply to author
Forward
0 new messages