bedGraph description error
3 views
Skip to first unread message
Stathis Kanterakis
Mar 2, 2018, 12:05:10 PM3/2/18
to gen...@soe.ucsc.edu
Hello and apologies if you get a million of these questions.
I'm referring to the bedGraph spec from here:
Specifically the following statements:
(1)
The chromosome coordinates are zero-based, half-open. This means that the first chromosome position is 0, and the last position in a chromosome of length N would be N - 1.
(2)
This example specifies 9 separate data points in three tracks on chr19 in the region 49,302,001 to 49,304,701.
...
chr19 49304400 49304700 1.00(1) It took me some time to parse this statement. Specifically I was confused thinking that an interval ending at N under 0-based, half-open, should be chr : X - N rather than chr : X - N-1. I see what you mean though because in a 0-based system, the coordinate N is N-1. IMHO the following statement would have been more helpful and clear:
The chromosome coordinates are zero-based, half-open. This means that an interval from 1 to N (inclusive) would be represented as 0 to N.
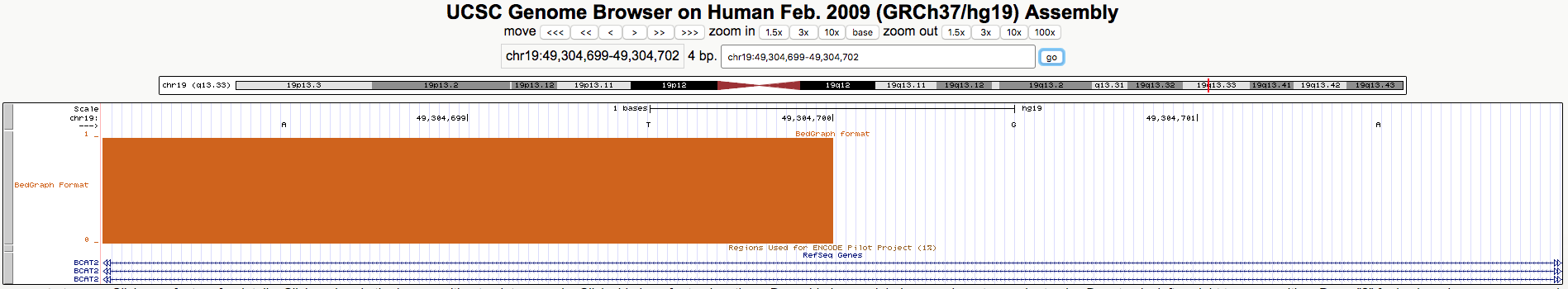
Then in statement (2) the 0-based interval chr19:49304400-49304700 supposedly includes coordinate 49304701, which is wrong. Here is a zoomed-in screenshot of your example that clearly shows that position 49304701 is not included.

I think what you meant to say is:
This example specifies 9 separate data points in three tracks on chr19 in the region 49,302,001 to 49,304,700.
I'm fairly confident about by 0 vs 1-based skills at this point in my career but this made be doubt again :P
Hopefully you can fix this so that others won't be confused.
Best,
Stathis
Reply all
Reply to author
Forward
0 new messages
