web BLAT not finding exons in repeat rich transcript
15 views
Skip to first unread message
Mike Gilchrist
Apr 28, 2016, 11:35:21 AM4/28/16
to gen...@soe.ucsc.edu
Dear UCSC Genome Informatics Group,
I have blatted a 756 bp, repeat rich, putative transcript sequence
against the Xenopus tropicalis genome.
The transcript has four exons spanning about 7kb of the genome.
BLAT reports ~200 matches to most (bases 57-756) of the first exon, but
fails to find the three downstream exons (which are ~not repetitive). By
chance(?) the first exon is repetitive up to ~ the 3' end, and the
'correct' match for this fragment is the first of those reported. The
three other exons are found easily by BLAT when searched with the
sequence absent the first exon.
The sequence(s) are pasted below:
I understand that repeats cause problems for BLAT (and BLAST, etc), but
it is very convenient for me to have a quick graphical web interface to
these searches as I work through a series of problematic gene models.
Is this an unfixable problem?
Thanks for your attention.
mike
>Xetro.K03375|Xetro.K03375.1 (exons 1-4)
CGATGCTCTCTCCCATATGTCCCCAATCCGGCTAATCTGTCCCTGCCTAACTGTAAACACCCAAAGGATCCAACACCAAAGGAAAAACAAAGCAATGTTG
TATACGCAGTCCAGTGTAGCGAGGAGTGCACAGACCTTTACATTGGTGAGACAAAGCAACTGCTCTCCAAGCGAATGGCTCCGCATAGGAGGGCGAACAC
TACAGGCCAGGACTCTGCAGTATTTCTACACCTAAAAGACAAGGGACACTCCTTTGAAAATAGCAATGTCCAACTTTTGGACAAAGAAGACCGCTGGTTT
GAAAGAGGTGTAAAAGAGGCCATTCATGTCAAAGTGGAGAAACCATCCCTAAACAGAGGCGGGGGACTTCGACACCATCTGTCTGCTACATACAATGCTG
TTCTAACATCTGTACCCCGGCAGTTTCAGAACTCTTCACACATCCATTCATGCAACTCTAACAAGAAATCAACTCCAAGTGAGCTTTTTTTATGTGAACT
GGATAACTGTGGAAAAGTATTCTCTAAGAGACAATATTTGAATTATCATCAGAAATACCAGCATGTGAATCAGCGTACCTTTTGCTGTCCAGTACCAGAG
TGTGGGAGAAAATTTAACTTCAAAAAACATTTAAAGGAACATGAAAAGAGGCACAGCGACCAACGAGACTTTATCTGTGAATTCTGTGCTCGTGCTTTCC
GTAGCAGCAGTAACCTCATCATCCACCACCGAATACACACTGGGGAGAAACCACTT
>Xetro.K03375|Xetro.K03375.1:466-756 (exons 2-4)
AAATCAACTCCAAGTGAGCTTTTTTTATGTGAACTGGATAACTGTGGAAAAGTATTCTCTAAGAGACAATATTTGAATTATCATCAGAAATACCAGCATG
TGAATCAGCGTACCTTTTGCTGTCCAGTACCAGAGTGTGGGAGAAAATTTAACTTCAAAAAACATTTAAAGGAACATGAAAAGAGGCACAGCGACCAACG
AGACTTTATCTGTGAATTCTGTGCTCGTGCTTTCCGTAGCAGCAGTAACCTCATCATCCACCACCGAATACACACTGGGGAGAAACCACTT
--
Mike Gilchrist
Group Leader
Vertebrate Systems Laboratory
The Francis Crick Institute
The Ridgeway
London, NW7 1AA
mike.gi...@crick.ac.uk
Tel: 0208 816 2451
Fax: 0208 906 4477
http://www.crick.ac.uk/mike-gilchrist
The Francis Crick Institute Limited is a registered charity in England and Wales no. 1140062 and a company registered in England and Wales no. 06885462, with its registered office at 215 Euston Road, London NW1 2BE.
I have blatted a 756 bp, repeat rich, putative transcript sequence
against the Xenopus tropicalis genome.
The transcript has four exons spanning about 7kb of the genome.
BLAT reports ~200 matches to most (bases 57-756) of the first exon, but
fails to find the three downstream exons (which are ~not repetitive). By
chance(?) the first exon is repetitive up to ~ the 3' end, and the
'correct' match for this fragment is the first of those reported. The
three other exons are found easily by BLAT when searched with the
sequence absent the first exon.
The sequence(s) are pasted below:
I understand that repeats cause problems for BLAT (and BLAST, etc), but
it is very convenient for me to have a quick graphical web interface to
these searches as I work through a series of problematic gene models.
Is this an unfixable problem?
Thanks for your attention.
mike
>Xetro.K03375|Xetro.K03375.1 (exons 1-4)
CGATGCTCTCTCCCATATGTCCCCAATCCGGCTAATCTGTCCCTGCCTAACTGTAAACACCCAAAGGATCCAACACCAAAGGAAAAACAAAGCAATGTTG
TATACGCAGTCCAGTGTAGCGAGGAGTGCACAGACCTTTACATTGGTGAGACAAAGCAACTGCTCTCCAAGCGAATGGCTCCGCATAGGAGGGCGAACAC
TACAGGCCAGGACTCTGCAGTATTTCTACACCTAAAAGACAAGGGACACTCCTTTGAAAATAGCAATGTCCAACTTTTGGACAAAGAAGACCGCTGGTTT
GAAAGAGGTGTAAAAGAGGCCATTCATGTCAAAGTGGAGAAACCATCCCTAAACAGAGGCGGGGGACTTCGACACCATCTGTCTGCTACATACAATGCTG
TTCTAACATCTGTACCCCGGCAGTTTCAGAACTCTTCACACATCCATTCATGCAACTCTAACAAGAAATCAACTCCAAGTGAGCTTTTTTTATGTGAACT
GGATAACTGTGGAAAAGTATTCTCTAAGAGACAATATTTGAATTATCATCAGAAATACCAGCATGTGAATCAGCGTACCTTTTGCTGTCCAGTACCAGAG
TGTGGGAGAAAATTTAACTTCAAAAAACATTTAAAGGAACATGAAAAGAGGCACAGCGACCAACGAGACTTTATCTGTGAATTCTGTGCTCGTGCTTTCC
GTAGCAGCAGTAACCTCATCATCCACCACCGAATACACACTGGGGAGAAACCACTT
>Xetro.K03375|Xetro.K03375.1:466-756 (exons 2-4)
AAATCAACTCCAAGTGAGCTTTTTTTATGTGAACTGGATAACTGTGGAAAAGTATTCTCTAAGAGACAATATTTGAATTATCATCAGAAATACCAGCATG
TGAATCAGCGTACCTTTTGCTGTCCAGTACCAGAGTGTGGGAGAAAATTTAACTTCAAAAAACATTTAAAGGAACATGAAAAGAGGCACAGCGACCAACG
AGACTTTATCTGTGAATTCTGTGCTCGTGCTTTCCGTAGCAGCAGTAACCTCATCATCCACCACCGAATACACACTGGGGAGAAACCACTT
--
Mike Gilchrist
Group Leader
Vertebrate Systems Laboratory
The Francis Crick Institute
The Ridgeway
London, NW7 1AA
mike.gi...@crick.ac.uk
Tel: 0208 816 2451
Fax: 0208 906 4477
http://www.crick.ac.uk/mike-gilchrist
The Francis Crick Institute Limited is a registered charity in England and Wales no. 1140062 and a company registered in England and Wales no. 06885462, with its registered office at 215 Euston Road, London NW1 2BE.
Cath Tyner
Apr 29, 2016, 2:45:27 PM4/29/16
to Mike Gilchrist, UCSC Genome Browser Public Help Forum
Hello Mike,
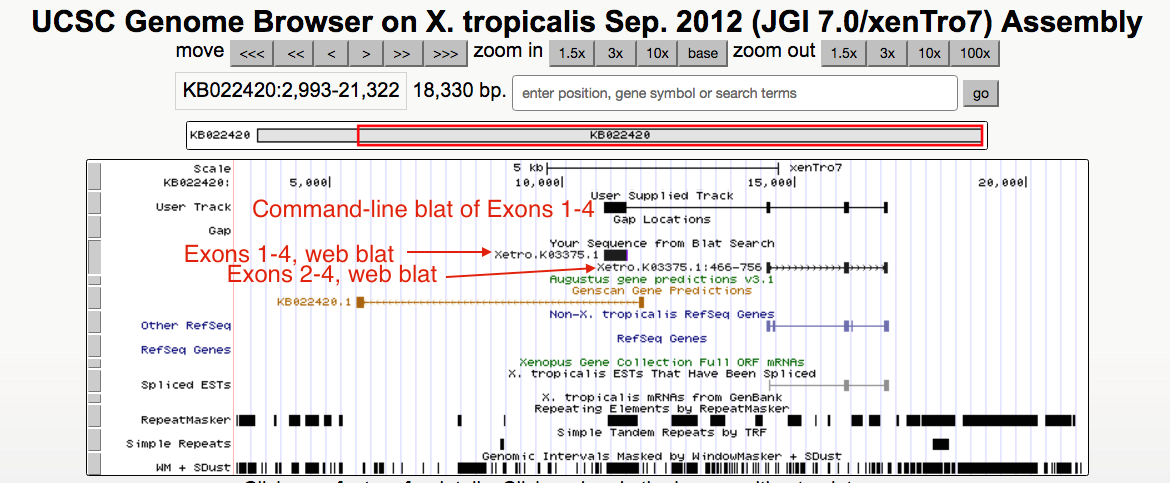
Thank you for using the UCSC Genome Browser and for submitting your question regarding web-based BLAT not finding expected regions. We are currently investigating the example that you have described as a potential issue to fix. In the meantime, you may want to consider using command-line BLAT to find repeat-rich sequence, even though it may not be as fast as the web tool.
Below is a screen shot of the command-line BLAT hit for KB022420 using the
For the command-line BLAT query, I used http://hgdownload.soe.ucsc.edu/goldenPath/xenTro2/bigZips/xenTro2.2bit with no special parameters:
blat xenTro7.2bit fasta1.fa output1.psl
I then uploaded the output for KB022420 as a custom track/bed file.
Resources:
http://hgdownload.soe.ucsc.edu/goldenPath/xenTro2/bigZips/ (xenTro2.2bit)
https://users.soe.ucsc.edu/~kent/src/
http://genome.ucsc.edu/goldenPath/help/blatSpec.html
http://hgdownload.soe.ucsc.edu/downloads.html#source_downloads
http://hgdownload.soe.ucsc.edu/admin/exe/ (bedToPsl)
Thank you for using the UCSC Genome Browser and for submitting your question regarding web-based BLAT not finding expected regions. We are currently investigating the example that you have described as a potential issue to fix. In the meantime, you may want to consider using command-line BLAT to find repeat-rich sequence, even though it may not be as fast as the web tool.
Below is a screen shot of the command-line BLAT hit for KB022420 using the
full
sequence (not the subset)
that you provided (for exons 1-4). You can see that all 4 exons were matched, compared to the web-based blat sequence below it.For the command-line BLAT query, I used http://hgdownload.soe.ucsc.edu/goldenPath/xenTro2/bigZips/xenTro2.2bit with no special parameters:
blat xenTro7.2bit fasta1.fa output1.psl
I then uploaded the output for KB022420 as a custom track/bed file.

Resources:
http://hgdownload.soe.ucsc.edu/goldenPath/xenTro2/bigZips/ (xenTro2.2bit)
https://users.soe.ucsc.edu/~kent/src/
http://genome.ucsc.edu/goldenPath/help/blatSpec.html
http://hgdownload.soe.ucsc.edu/downloads.html#source_downloads
http://hgdownload.soe.ucsc.edu/admin/exe/ (bedToPsl)
Thank you again for your inquiry and for using the UCSC Genome Browser.
Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private data help: Email
genom...@soe.ucsc.eduEnjoy,
Cath
. . .
Cath Tyner. . .
UCSC Genome Browser, Software QA & User Support
UC Santa Cruz Genomics Institute
--
--- You received this message because you are subscribed to the Google Groups "UCSC Genome Browser discussion list" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
Cath Tyner
May 2, 2016, 4:42:10 PM5/2/16
to Mike Gilchrist, UCSC Genome Browser Public Help Forum
Thanks for the response, Mike. We greatly appreciate questions and feedback from all of our UCSC Genome Browser users. You are correct, we compromise accuracy in certain situations (e.g., rich repeats) for speed in the web-based blat tool. It's a very common question, as outlined in our FAQ:
https://genome.ucsc.edu/FAQ/FAQblat.html#blat1b
As you suggested, having an option to run web-based blat with various parameters (e.g., "slower but better") would be ideal in some situations. Your suggestion has been noted for review.
To keep alert of future data and software changes, you can sign up for the UCSC Genome Browser announcements list:
Subscribe: Email genome-annou...@soe.ucsc.edu
Unsubscribe: Email genome-announ...@soe.ucsc.edu
https://genome.ucsc.edu/FAQ/FAQblat.html#blat1b
As you suggested, having an option to run web-based blat with various parameters (e.g., "slower but better") would be ideal in some situations. Your suggestion has been noted for review.
To keep alert of future data and software changes, you can sign up for the UCSC Genome Browser announcements list:
Subscribe: Email genome-annou...@soe.ucsc.edu
Unsubscribe: Email genome-announ...@soe.ucsc.edu
Thank you again for your inquiry and for using the UCSC Genome Browser.
Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private data help: Email
genom...@soe.ucsc.eduOn Sat, Apr 30, 2016 at 4:41 AM, Mike Gilchrist <mike.gi...@crick.ac.uk> wrote:
Thanks - good to know it was worth sending in.
One of the reasons why I like the browser - you guys listen and fix stuff.
So 'obviously' you should run the web BLAT with the same settings as the standalone BLAT!
I guess it's suboptimal for something else - speed?
Maybe a selectable option 'slow but better' on the web site?
Thanks again,
mike
Mike Gilchrist
Apr 16, 2018, 4:47:54 PM4/16/18
to gen...@soe.ucsc.edu
This is a follow up to a discussion with Cath Tyner in 2016.
It's about web BLAT missing an exon alignment under some conditions, but not others.
This time there's no obvious repetitve sequence involved.
Once again, (part of) a Xenopus tropicalis exon is not found by web BLAT under some condition, but is found under others.
It's the 20 bp start to the ORF, which is part of the first exon of the gene.
Here's the inconsistent bit:
- BLAT finds this part of the exon when searching with <= 1702 bp of the ORF from the ATG.
- but fails to find it with > 1702 bp - all sequences starting from the same position (the ATG)
See pasted-in graphic below, and sequences attached.
If there's a rational explanation for this behaviour I'd love to know, but it would be nice if it didn't do this.
Interestingly on our (slightly older) installation of your browser, it fails on all sequences...
Best wishes,
Mike Gilchrist

It's about web BLAT missing an exon alignment under some conditions, but not others.
This time there's no obvious repetitve sequence involved.
Once again, (part of) a Xenopus tropicalis exon is not found by web BLAT under some condition, but is found under others.
It's the 20 bp start to the ORF, which is part of the first exon of the gene.
Here's the inconsistent bit:
- BLAT finds this part of the exon when searching with <= 1702 bp of the ORF from the ATG.
- but fails to find it with > 1702 bp - all sequences starting from the same position (the ATG)
See pasted-in graphic below, and sequences attached.
If there's a rational explanation for this behaviour I'd love to know, but it would be nice if it didn't do this.
Interestingly on our (slightly older) installation of your browser, it fails on all sequences...
Best wishes,
Mike Gilchrist

On 02/05/2016 21:42, Cath Tyner wrote:
Thanks for the response, Mike. We greatly appreciate questions and feedback from all of our UCSC Genome Browser users. You are correct, we compromise accuracy in certain situations (e.g., rich repeats) for speed in the web-based blat tool. It's a very common question, as outlined in our FAQ:
https://genome.ucsc.edu/FAQ/FAQblat.html#blat1b
As you suggested, having an option to run web-based blat with various parameters (e.g., "slower but better") would be ideal in some situations. Your suggestion has been noted for review.
To keep alert of future data and software changes, you can sign up for the UCSC Genome Browser announcements list:
Subscribe: Email genome-annou...@soe.ucsc.edu
Unsubscribe: Email genome-announ...@soe.ucsc.edu
Thank you again for your inquiry and for using the UCSC Genome Browser.Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private data help: Emailgenom...@soe.ucsc.edu
CathCath Tyner
. . .
UCSC Genome Browser, Software QA & User SupportUC Santa Cruz Genomics Institute
--
Mike Gilchrist Group Leader Vertebrate Systems Laboratory The Francis Crick Institute The Ridgeway London, NW7 1AA mike.gi...@crick.ac.uk
Tel: 0203 7962 418 http://www.crick.ac.uk/mike-gilchrist
The Francis Crick Institute Limited is a registered charity in England and Wales no. 1140062 and a company registered in England and Wales no. 06885462, with its registered office at 1 Midland Road London NW1 1AT
Brian Lee
Apr 17, 2018, 5:08:11 PM4/17/18
to Mike Gilchrist, UCSC Genome Browser Mailing List
Dear Mike,
Thank you for using the UCSC Genome Browser and your question about how BLAT alignments work.
The short response to the question is that sensitivity to BLAT queries is just in the range of 20 basepairs, and this first exon is quite small at just about this limit of detection.
As the algorithm behind BLAT is attempting to find the best match for the entire sequence entered and the sequence entered increases in size with more matching bases further away there is likely an increased penalty in the scoring to include a massive gap to jump over and catch that small 20-base span from the start of the query.
It is frustrating that extending the last exon can make BLAT miss that first small exon, which would both give the align full-query span and start at ATG, but if there were more bases in the first exon (beyond the near 20 bp detection limit), say for example by increasing the first sequence by 15 more bases to 35bp, then the algorithm would have an increased possibility to include that sequence with high confidence. Below is an example query where a few added bases allow a match:
>tag-example15morebasesOnFirstMatchBeforeATG
TGCAGCCAGAACACCATGACACAAGACTACGACAACAAACGGCCGGTGTTGGTGCTTCAGAACGATGGGCTGTACCAGCAGAGGAGATCCTACACCAGTGAGGATGAAGCCTGGAAGTCCTTTCTTGAAAACCCCCTCACGGCGGCCACCAAAGCGATGATGAGCATAAATGGAGATGAAGACAGTGCAGCCGCCTTAGGCCTTTTATATGATTATTACAAGGTTCCCAGGGAAAGGAGGCCGTCGGCAGCAAAACAGGAGCATGATCATGCGGATCAGGAGCACAACAAAAGGAATGGTTTGCCTCAGATCAATGAACAAGCCCTGCTTTCAGAGAACAGAGTGCAGGTACTGAAAACAGTGCCCTTTAACATTGTGGTACCCCTGGCCAACCAGGTGGATAAAAGAGGTCACCTGACCACGCCAGACACTACAGCCGCAGTCTCTATTACGTCACTACCGGCTCATCCCATCAAAACTGAATCCCAGAGCCATTGCTTCTCTGTGGGCCTTCAGAGTGTGTTCCACACGGAACCTACGGAAAGGATTGTCACCTTTGATCGAACTGTCCCTTCTGACCACTTCACATCTAACAGCCAGCCACCTAACTCCCAGAGGCGCACCCCAGACTCCACATTCTCTGAGACATACAAGGAAGATGTTCCAGAGGTTTTCTTTCCACCTGACCTGAGCCTACGAATGGGCAGCATGAATTCTGAAGACTATGTTTTCGATTCTGTTGCTGGGAATAATTTTGAATATACCCTTGAGGCATCAAAATCCCTTAGGCCTAAACCTGGGGACAGCACTATGACGTATCTGAACAAAGGCCAGTTTTACCCTATAACCCTAAAGGAGATTGGCGGCAACAAAGGAATACACCATCCAATCAGTAAAGTCCGGAGTGTGATTATGGTTGTGTTTGCTGATGACAAAAGCAGAGAGGACCAGCTCCGCCATTGGAAGTACTGGCATTCACGGCAGCACACAGCAAAGCAGCGATGCATCGACATAGCCGACTACAAAGAGAGCTTTAACACCATCAGTAACATTGAGGAGATTGCGTACAACGCCATTTCTTTTACCTGGGACCTGAACGATGAAGGAAAGGTGTTTATATCTGTGAATTGCCTGAGTACAGACTTCTCCTCCCAGAAAGGAGTGAAAGGGCTGCCGCTGAACCTCCAGATTGACACGTACAGCTATAACAACAGGAGTAACAAGCCTGTGCACAGAGCCTACTGCCAGATTAAAGTATTCTGTGACAAGGGGGCAGAACGTAAAATAAGAGACGAGGAACGCAAACAGAGCAAGCGAAAAGTCCAGGATGTTAAGGTTGGGCTTCCTCCATCTCACAAGAGGACAGATATCACTGTGTTTAAGCCTATGATGGATCTGGACACTCAGCCAGTCCTGTTTATCCCAGATGTGCATTTTGCCAACCTCCAACGCACGACACATGTTCTTCCCATAGCACCTGAGGACATGGAAGGAGAATTGAGCCCCGGAATGAAGAGAGTGCCCTTCTCCCCCGAAGAGGATTTCACTGCACCCCCTGCTAAGCTGCCCCGGGTGGATGAACCAAAAAGAGTTTTGCTGTATGTCAGGAGGGAGACAGAGGAAGTTTTTGATGCTCTAATGCTCAAGACACCAACACTGAAAGGGTTAATGGAGGCTGTCTCTGACAAATATGAAGTCCCCATTGAAAAAATTGGAAAGA
In essence it is the very small 20bp first exon that is just at the limit of detection that gives a low confidence for BLAT to include the results as the increased size of searches moves farther away from that first exon and decreases support for an inclusion to that location across such a wide gap.
Thank you again for your inquiry and for using the UCSC Genome Browser. If you have any further questions and reply to gen...@soe.ucsc.edu messages will be archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
All the best,
Brian Lee
UC Santa Cruz Genomics Institute
Training videos & resources: http://genome.ucsc.edu/training/index.html
On Mon, Apr 16, 2018 at 10:02 AM, Mike Gilchrist <mike.gi...@crick.ac.uk> wrote:
This is a follow up to a discussion with Cath Tyner in 2016.
It's about web BLAT missing an exon alignment under some conditions, but not others.
This time there's no obvious repetitve sequence involved.
Once again, (part of) a Xenopus tropicalis exon is not found by web BLAT under some condition, but is found under others.
It's the 20 bp start to the ORF, which is part of the first exon of the gene.
Here's the inconsistent bit:
- BLAT finds this part of the exon when searching with <= 1702 bp of the ORF from the ATG.
- but fails to find it with > 1702 bp - all sequences starting from the same position (the ATG)
See pasted-in graphic below, and sequences attached.
If there's a rational explanation for this behaviour I'd love to know, but it would be nice if it didn't do this.
Interestingly on our (slightly older) installation of your browser, it fails on all sequences...
Best wishes,
Mike Gilchrist
On 02/05/2016 21:42, Cath Tyner wrote:
Thanks for the response, Mike. We greatly appreciate questions and feedback from all of our UCSC Genome Browser users. You are correct, we compromise accuracy in certain situations (e.g., rich repeats) for speed in the web-based blat tool. It's a very common question, as outlined in our FAQ:
https://genome.ucsc.edu/FAQ/FAQblat.html#blat1b
As you suggested, having an option to run web-based blat with various parameters (e.g., "slower but better") would be ideal in some situations. Your suggestion has been noted for review.
To keep alert of future data and software changes, you can sign up for the UCSC Genome Browser announcements list:
Subscribe: Email genome-announce+subscribe@soe.ucsc.edu
Unsubscribe: Email genome-announce+unsubscribe@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/c1a0e298-4b02-73d3-9127-334e72957d2b%40crick.ac.uk.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
Reply all
Reply to author
Forward
0 new messages
