bigBed file behaves differently in a hub than as a custom_track??? no gene names!
Hendrickson, Curtis (Campus)
Dear TrackHub folks
Here’s a session and a hub for the question below:
https://data.genome.uab.edu/ccts/bmi/ics/ross,s/track_hub/
It contains the de novo contigs from a viral genome sequence, some gene annotations (bed/bigBed) and a set of mapped reads (bam).
I have a problem with the bigBed display, and a question about configuring default BAM file display.
We created some bigBed files that show gene annotations mapped from similar strains via Exonerate.
When I load these as “custom_tracks” and view them in “full” mode, they nicely display their gene names (column 4 of bed file)
However, when I use the same .bb file inside by track_hub, to comes up grey, with no names at all, and the introns are not displayed, all of which is quite dismaying. I figure I’ve messed up some configurations in the trackDb.txt file, but haven’t been able to figure out what I’ve done wrong.
Hub/BigBed problem screen shot:
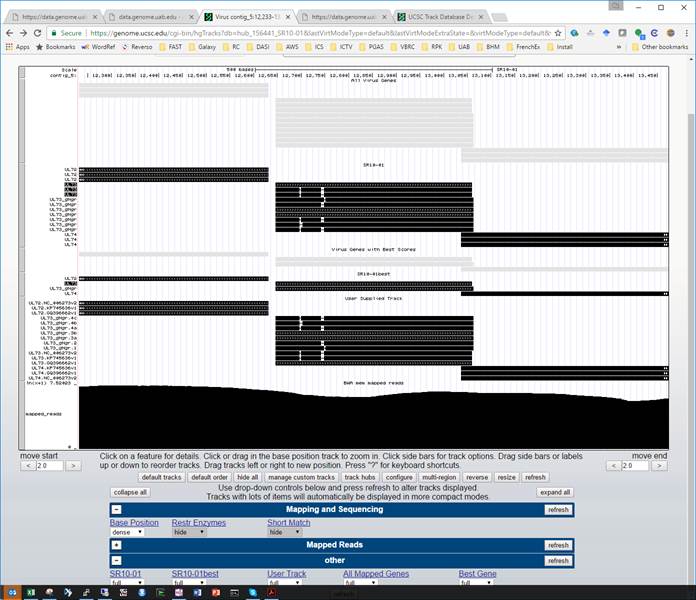
BAM file question:
In this session I’ve set the bamfile as a bargraph, but I can’t figure out how to specify in trackDb.txt that it should default to that display mode. I don’t see any tags for that at https://genome.ucsc.edu/goldenPath/help/trackDb/trackDbHub.html
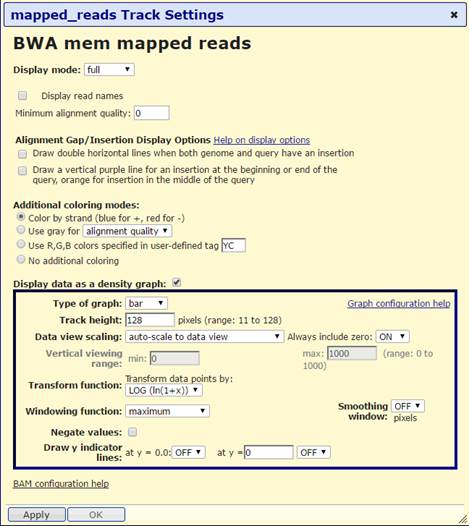
Regards,
Curtis
PS _ thanks again for your great help in the past!
Cath Tyner
longLabel All Virus Genes
useScore 1
type bigBed
* Unsubscribe: Email genome-announce+unsubscribe@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/40992bc27af94fb98058fc7d73cd08db%40uabex8.ad.uab.edu.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
