Re: Using links in bigBed extra feilds
19 views
Skip to first unread message
Mitchell Vollger
Oct 19, 2017, 11:34:55 AM10/19/17
to gen...@soe.ucsc.edu
Never mind, please disregard my previous email.
On Wed, Oct 18, 2017 at 5:39 PM, Mitchell Vollger <mvol...@uw.edu> wrote:
Hello,I am making my own track hub and I have a rather particular use case. I am displaying my data with bigBed9+2, so the 9 bed fields and two extra fields. The .as file file that describes my data looks like this:table CHM1_ABP"A collapse from CHM1 assembled by ABP"(string chrom; "Reference sequence chromosome or scaffold"uint chromStart; "Start position of feature on chromosome"uint chromEnd; "End position of feature on chromosome"string name; "Name of gene"uint score; "Score"char[1] strand; "+ or - for strand"uint thickStart; "Coding region start"uint thickEnd; "Coding region end"uint reserved; "Green on + strand, Red on - strand"string ID; "Correlation Clustering ID"lstring html; "Description of CC results")I have this successfully working and displayed on the genome browser, except for one thing. The last entry in my bed file is actually a long html. This html string includes several links, all of which work when I load the html string on its own. But when I click on a region in the genome browser to bring up the description and that html, the links are not active, they just appears as plain text.Is there anyway I can encode active hyperlinks into a bigBed file?Thanks!Mitchell
Mitchell Vollger
Oct 19, 2017, 11:35:10 AM10/19/17
to gen...@soe.ucsc.edu
Brian Lee
Oct 19, 2017, 12:25:04 PM10/19/17
to Mitchell Vollger, gen...@soe.ucsc.edu
Dear Mitchell,
Thank you for your message and using the UCSC Genome Browser to build track hubs.
We also received your message asking to disregard this question, and we are glad that you have resolved the issue. For other users that may be searching for this question in the future, we would like to go ahead and post details to a solution for their future reference.
Here is a temporary small hub that illustrates adding a 13th column and using that column to build a URL to an external site that might be of interest. Here is a link to the trackDb.txt
Here is a link to load the hub: http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chrX%3A160768013-160799663&hubUrl=http://hgwdev.cse.ucsc.edu/~brianlee/hubTesting/tempIdeas/newHub/hub.txt
If you click this item you will see there is now a displayed link to MGI ID for item.
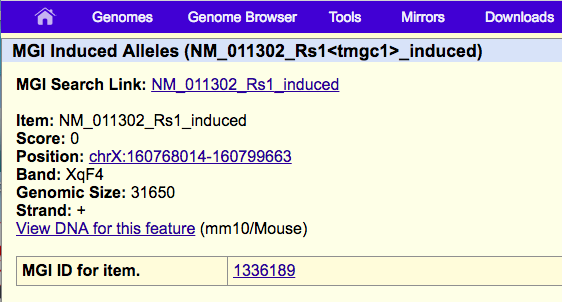
This will take you to http://www.informatics.jax.org/marker/MGI:1336189
This was done by adding a 13th column to some bed data (where since the exact mapping was unknown 1336189 was added for all items), for example like this:
chrX 160768013 160799663 NM_011302_Rs1<tmgc1>_induced 0 + 160768013 160799663 0 6 224,25,105,141,195,5144, 0,11345,13792,22761,23379,26506, 1336189
This bed was made into a bigBed with this autosql definition file where I added an additional column called MGI_ID:
Built into a bigBed with this command:
bedToBigBed -type=bed12+ Alleles_Induced.13.bed -as=bed13.as http://hgdownload.cse.ucsc.edu/goldenPath/mm10/bigZips/mm10.chrom.sizes Alleles_Induced.13.bb
Then in the trackDb.txt this additional urls line was added (but if there were multiple additional columns, many URLs could be built):
urls MGI_ID="http://www.informatics.jax.org/marker/MGI:$$"
There is also a link at the top in these detail pages that uses the "name" field in the bed that is made into a link with this trackDb.txt line:
You can add many additional columns and add many additional links if you desire to your data. There are also other options to add searchIndexes on the names and searchTrix files that further allow increased search options if desired.
You can learn more about the url, urls, searchIndex and searchTrix fields here:
Thank you again for your inquiry and using the UCSC Genome Browser. If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
All the best,
Brian Lee
UC Santa Cruz Genomics Institute
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CAHPLNqE65HhOQA3r3hv%2BCUmFmqOMcNFEX7Z2PsPrAyqiB%2Bvkvg%40mail.gmail.com.--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
Reply all
Reply to author
Forward
0 new messages
