inconsistency between genome browser and table browser
Yin, Lihui
Dear UCSC genome browser team,
Recently I used the table browser for the RHCE gene (NM_020485). However the exon position isn’t consistent with what I got from the genome browser. Would you please help check for me anything I didn’t do correctly?
In the genome browser, the exon 1 ends at chr1:25747130, which is consistent with the result calculated from NCBI (Go to:
LOCUS NG_009208 74944 bp DNA linear PRI 22-MAY-2015, /transcript_id="NM_020485.4")
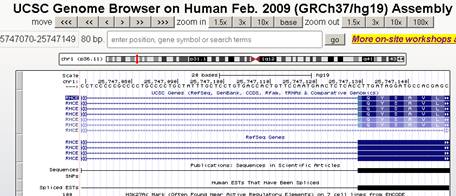
In the table browser, the assembly is Feb. 2009 (hg19)
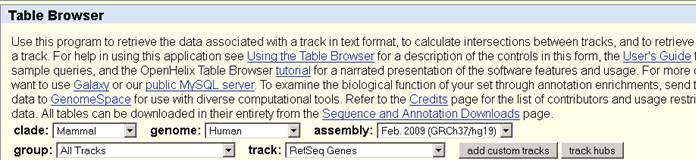
The transcript number is NM_020485
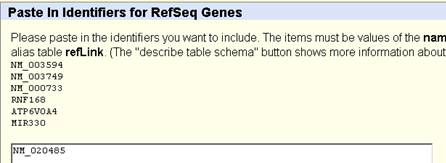
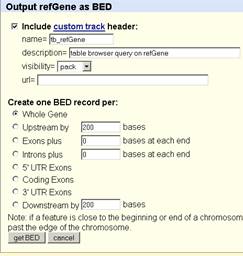
In the BED file, the exon length and start position were showed below:
track name="tb_refGene" description="table browser query on refGene" visibility=3 url= | |||||||||||||
chr1 | 25688739 | 25747363 | NM_020485 | 0 | - | 25689017 | 25747277 | 0 | 10 | 305,74,80,134,138,167,148,151,187,234, | 0,8218,13100,23462,26727,28500,29745,40347,46434,58390, | ||
Using this table, it was calculated that the chromosome coordinate for exon 1 is 25747363-25747059 (- strand), different from the genome browser finding.
Could you think of anything that causes this or I might go wrong?
Appreciate your help!
Lihui
-- ******************** CONFIDENTIALITY NOTICE ******************** This transmission may contain confidential information. If you have received this transmission in error, please notify the sender immediately and destroy this material.
