adding a genome using a custom track hub - are pipe's allowed in chr names?
Curtis Hendrickson (Campus)
Dear UCSC,
We are working on creating a track hub for the virus Human Herpesvirus 5, based initially on the NCBI RefSeq entry NC_006273.2.
ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses/Human_herpesvirus_5_uid14559/
this organism has a single segment genome (1 chromosome).
However, the fasta file provided by NCBI RefSeq, gives a “chromosome name” of
>gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
For compatibility with RefSeq, we created our hub .2bit file and .bigbed files using that same “chromosome” name, so that VCF files created by users who download their reference genome from RefSeq will work.
https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_hub/hub.txt
This hub passes muster with hubCheck (see below).
However, that seems to be causing some indigestion in the UCSC Browser.
I can get the display to come up, including the transcript track we provide, but every time we try and interact, it hangs.
Also, the only way to get to a display is to go through the “(sequences)” link.
Most other pathways in end in an error window:
Warning/Error(s):
- Sorry, couldn't locate gi|155573622|ref|NC_006273.2|:1-235,646 in genome database
OK
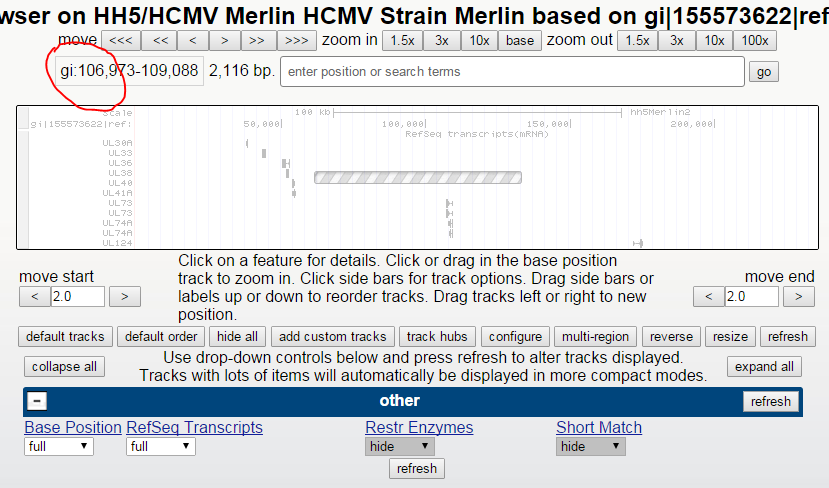
And attempts to zoom, show an chromosome name that is truncated after the “gi” – ie at the first pipe (see figure below).
Questions
1. Is our guess correct that the pipes (|) in the chromosome name are at fault?
2. Do you support some sort of “aliases” file like IGV does? (We use that heavily to allow us to use “nicer” chromosome names, while still being compatible with things computed with RefSeq acquired reference fastas)
Thanks!
Regards,
Curtis
Research Associate, Informatics Unit
Center for Clinical and Translational Science
University of Alabama at Birmingham
Tel: 205.975.5240
Email: cur...@uab.edu
# hub with |’s in chromosome name passes hubCheck
[curtish@cheaha tmp]$ rm -rf hubCheck ;(module load ngs-ccts/ucsc_kent/2014-03-05 ; hubCheck -verbose=2 -udcDir=hubCheck https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt)
udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt, hubCheck)
checking http remote info on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
bitmap file hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt/bitmap does not already exist, creating.
reading http/https/ftp data - 258 bytes at 0 - on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt, hubCheck)
checking http remote info on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt
bitmap file hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt/bitmap does not already exist, creating.
reading http/https/ftp data - 406 bytes at 0 - on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt
hub https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
shortLabel hh5Merlin2
longLabel Track hub for HCMV Strain Merlin based on gi|155573622|ref|NC_006273.2|
genomes.txt has 1 elements
udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/trackDb.txt, hubCheck)
checking http remote info on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/trackDb.txt
bitmap file hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/trackDb.txt/bitmap does not already exist, creating.
reading http/https/ftp data - 143 bytes at 0 - on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/trackDb.txt
checking _hh5Merlin2._hh5Merlin2_refseq_mrna type bigBed 12 at https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273.2.mrna.bb
udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273.2.mrna.bb, hubCheck)
checking http remote info on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273.2.mrna.bb
bitmap file hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273.2.mrna.bb/bitmap does not already exist, creating.
reading http/https/ftp data - 8192 bytes at 0 - on https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273.2.mrna.bb
[curtish@cheaha tmp]$
View after clicking on (Sequences)
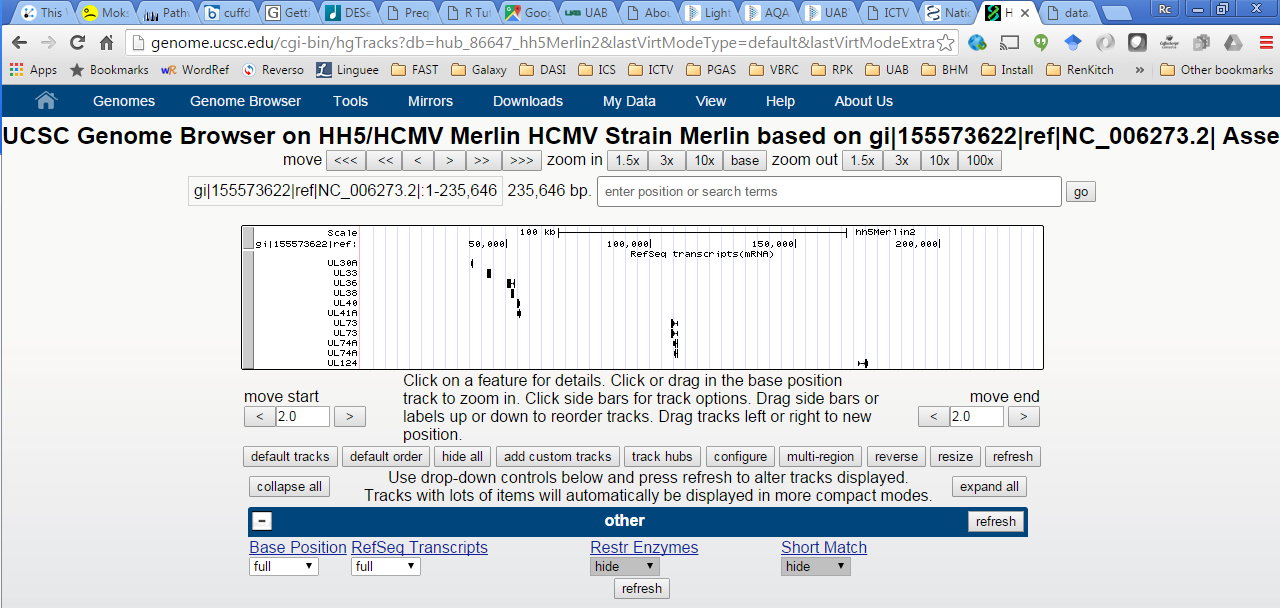
Figure: zoom in to UL73 (right click on UL73 transcript, select “zoom to…”)
Browser hangs trying to zoom….
Matthew Speir
Thank you for your question about including pipes in chromosome names in the UCSC Genome Browser.
As you have so far deduced, these pipes, or "|", in the chromosome names are not allowed. In the Genome Browser, your chromosome names should be all alpha or numeric characters, with the first character being a letter.
You suggestion of an alias file is excellent and I have made a note of it in our internal tracking system, however, I can't give an estimate of when this might be implemented.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible Google Groups forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
Matthew Speir
UCSC Genome Bioinformatics Group
--
Hiram Clawson
I would recommend using the sequence ID NC_006273.2 for the 'chromosome' name, for example:
http://genome-preview.cse.ucsc.edu/cgi-bin/hgTracks?hubUrl=http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/02/hub.ncbi.txt&genome=GCF_000845245.1_ViralProj14559&position=NC_006273.2
--Hiram
>>
>> Most other pathways in end in an error window:
>>
>>
>> * Sorry, couldn't locate gi|155573622|ref|NC_006273.2|:1-235,646 in genome
>>
>> OK
>>
>> And attempts to zoom, show an chromosome name that is truncated after the “gi”
>> – ie at the first pipe (see figure below).
>>
>>
>> _Questions_
>>
>> 1.Is our guess correct that the pipes (|) in the chromosome name are at fault?
>>
>> 2.Do you support some sort of “aliases” file like IGV does? (We use that
>> compatible with things computed with RefSeq acquired reference fastas)
>>
>> Thanks!
>>
>> Regards,
>>
>> Curtis
>>
>> Research Associate, Informatics Unit
>>
>> Center for Clinical and Translational Science
>>
>> http://bioinformatics.uab.edu
>>
>> University of Alabama at Birmingham
>>
>> Tel: 205.975.5240
>>
>>
>> *_# hub with |’s in chromosome name passes hubCheck_*
>>
>> cid:image0...@01D18FFD.6F364320
>>
>> *_Figure: zoom in to UL73 (right click on UL73 transcript, select “zoom to…”)_*
>> Browser hangs trying to zoom….
>>
>>
>> --
>>
>
> --
>
Hiram Clawson
The assembly hubs are my project. It is a prototype work in progress.
I have automated the sequence of events from an NCBI download directory to
the files required for an assembly hub. We are considering how to make it
a viable public resource, there are difficulties. A rough copy of the
scripts that perform this process can be found in our source tree:
http://genome-source.cse.ucsc.edu/gitweb/?p=kent.git;a=tree;f=src/hg/utils/automation/genbank
Index to all assemblies:
http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/
http://genome-preview.cse.ucsc.edu/gbdb/hubs/genbank/
--Hiram
On 4/6/16 2:42 PM, Curtis Hendrickson (Campus) wrote:
> Hiram
>
>
>
> Thank you very much!
>
> That was, indeed, our fallback plan, to use the accession only, especially with gi numbers phasing out.
>
>
>
> However, from your URL, it looks like someone has already done a similar effort.
>
> Can you point me to some reference for this project - looks like someone did/is doing all the RefSeq genomes in a systematic way?
>
> http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/viral.ncbi.html
>
> so the HH5/HCMV stub is
>
> http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/02/GCF_000845245.1_ViralProj14559/GCF_000845245.1_ViralProj14559.description.html
>
>
>
> What is the status / roadmap for this project?
>
> How is the Genbank -> Bed file translation done for these?
>
> We had looked around for Genbank-> Bed file translators that would preserve CDS and exon structure, and hadn’t found anything we liked. Obviously we missed one.
>
>
>
> Regards,
>
> Curtis
>
>
> -----Original Message-----
> From: Hiram Clawson [mailto:hi...@soe.ucsc.edu]
> Sent: Wednesday, April 06, 2016 4:28 PM
> To: Curtis Hendrickson (Campus)
> Cc: gen...@soe.ucsc.edu; Blair Delane Heater; Elliot J Lefkowitz
> Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
>
>
>
> Good Afternoon Curtis:
>
>
>
> I would recommend using the sequence ID NC_006273.2 for the 'chromosome' name, for example:
>
>
>
> http://genome-preview.cse.ucsc.edu/cgi-bin/hgTracks?hubUrl=http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/02/hub.ncbi.txt&genome=GCF_000845245.1_ViralProj14559&position=NC_006273.2
>
>
>
> --Hiram
>
>
>
> On 4/6/16 2:21 PM, Matthew Speir wrote:
>
>> Hi Curtis,
>
>>
>
>> Thank you for your question about including pipes in chromosome names
>
>> in the UCSC Genome Browser.
>
>>
>
>> As you have so far deduced, these pipes, or "|", in the chromosome
>
>> names are not allowed. In the Genome Browser, your chromosome names
>
>> should be all alpha or numeric characters, with the first character being a letter.
>
>>
>
>> You suggestion of an alias file is excellent and I have made a note of
>
>> it in our internal tracking system, however, I can't give an estimate
>
>> of when this might be implemented.
>
>>
>
>> I hope this is helpful. If you have any further questions, please
>
>> archived on a publicly-accessible Google Groups forum. If your
>
>>>
>
>>> *_# hub with |’s in chromosome name passes hubCheck_*
>
>>>
>
>>> [curtish@cheaha tmp]$ rm -rf hubCheck ;(module load
>
>>> ngs-ccts/ucsc_kent/2014-03-05 ; hubCheck -verbose=2 -udcDir=hubCheck
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt)
>
>>>
>
>>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>
>>> ub/hub.txt,
>
>>> hubCheck)
>
>>>
>
>>> checking http remote info on
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
>
>>>
>
>>> bitmap file
>
>>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hu
>
>>> b.txt/bitmap
>
>>> does not already exist, creating.
>
>>>
>
>>> reading http/https/ftp data - 258 bytes at 0 - on
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
>
>>>
>
>>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>
>>> ub/genomes.txt,
>
>>> hubCheck)
>
>>>
>
>>> checking http remote info on
>
>
>>> xt
>
>>>
>
>>> bitmap file
>
>>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/ge
>>> nomes.txt/bitmap
>
>>> does not already exist, creating.
>
>>>
>
>>> reading http/https/ftp data - 406 bytes at 0 - on
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.t
>
>>> xt
>
>>>
>
>>> hub
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
>
>>>
>
>>> shortLabel hh5Merlin2
>
>>>
>
>>> longLabel Track hub for HCMV Strain Merlin based on
>
>>> gi|155573622|ref|NC_006273.2|
>
>>>
>
>>> genomes.txt has 1 elements
>
>>>
>
>>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>
>>> ub/hh5Merlin2/trackDb.txt,
>
>
>>>
>
>>> checking http remote info on
>
>
>>> 2/trackDb.txt
>
>>>
>
>>> bitmap file
>
>>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh
>>> 5Merlin2/trackDb.txt/bitmap
>
>>> does not already exist, creating.
>
>>>
>
>>> reading http/https/ftp data - 143 bytes at 0 - on
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin
>
>>> 2/trackDb.txt
>
>>>
>
>>> checking _hh5Merlin2._hh5Merlin2_refseq_mrna type bigBed 12 at
>
>>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin
>
>>> 2/NC_006273.2.mrna.bb
>
>>>
>
>>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>
>>> ub/hh5Merlin2/NC_006273.2.mrna.bb,
>
>
>>>
>
>>> checking http remote info on
>
>
>>> 2/NC_006273.2.mrna.bb
>
>>>
>
>>> bitmap file
>
>>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh
Curtis Hendrickson (Campus)
Hiram
Thank you very much!
That was, indeed, our fallback plan, to use the accession only, especially with gi numbers phasing out.
However, from your URL, it looks like someone has already done a similar effort.
Can you point me to some reference for this project - looks like someone did/is doing all the RefSeq genomes in a systematic way?
http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/viral.ncbi.html
so the HH5/HCMV stub is
What is the status / roadmap for this project?
How is the Genbank -> Bed file translation done for these?
We had looked around for Genbank-> Bed file translators that would preserve CDS and exon structure, and hadn’t found anything we liked. Obviously we missed one.
Regards,
Curtis
-----Original Message-----
From: Hiram Clawson [mailto:hi...@soe.ucsc.edu]
Sent: Wednesday, April 06, 2016 4:28 PM
To: Curtis Hendrickson (Campus)
Cc: gen...@soe.ucsc.edu; Blair Delane Heater; Elliot J Lefkowitz
Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
Good Afternoon Curtis:
>> xt
>>
>> bitmap file
>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/ge
>> nomes.txt/bitmap
>> does not already exist, creating.
>>
>> reading http/https/ftp data - 406 bytes at 0 - on
>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.t
>> xt
>>
>> hub
>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
>>
>> shortLabel hh5Merlin2
>>
>> longLabel Track hub for HCMV Strain Merlin based on
>> gi|155573622|ref|NC_006273.2|
>>
>> genomes.txt has 1 elements
>>
>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>> ub/hh5Merlin2/trackDb.txt,
>> hubCheck)
>>
>> checking http remote info on
>> 2/trackDb.txt
>>
>> bitmap file
>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh
>> 5Merlin2/trackDb.txt/bitmap
>> does not already exist, creating.
>>
>> reading http/https/ftp data - 143 bytes at 0 - on
>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin
>> 2/trackDb.txt
>>
>> checking _hh5Merlin2._hh5Merlin2_refseq_mrna type bigBed 12 at
>> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin
>> 2/NC_006273.2.mrna.bb
>>
>> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_p
>> ub/hh5Merlin2/NC_006273.2.mrna.bb,
>> hubCheck)
>>
>> checking http remote info on
>> 2/NC_006273.2.mrna.bb
>>
>> bitmap file
>> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh
>> 5Merlin2/NC_006273.2.mrna.bb/bitmap
>> does not already exist, creating.
Blair Delane Heater
Dear Matthew Speir,
Thanks for your help resolving the issue of the unsupported pipes. We attempted to configure our hh5Merlin2 track hub to show up under the Viruses option in the group pull down menu on the Genome Browser Gateway. If we changed the shortLabel in hub.txt to any text other than ‘Viruses’, it worked and appeared in the drop down menu. But, when the shortLabel was set to ‘Viruses’, disconnecting and reconnecting the track hub didn’t give error but reverted to the shortLabel of previously connected hub. For the moment, we set the shortLabel to be Viruses-HH5(HCMV) as seen below to associate the track hub with Viruses, while we can’t successfully place in within that option. Is it possible to set up the track hub under the ‘Viruses’ option in the group pull down menu? If so, what steps are we missing? If not, is there any standard for group names?

Thank you,
Blair Heater
From: Matthew Speir
Sent: Wednesday, April 6, 2016 4:22 PM
To: Curtis Hendrickson (Campus);
gen...@soe.ucsc.edu
Cc: Blair Delane Heater;
Elliot J Lefkowitz
Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
Curtis Hendrickson (Campus)
Matthew & Hiram,
A second question occurred to us after the problem with the pipes (|) in the chromosome names: Is there a problem with using period (.) in chromosome names that we have yet to trip over ?
In our initial testing with chrom=”NC_006273.2”, things seem to work well.
We noticed that Hiram has converted RefSeq/NCBI’s “NC_006273.2” to “NC_006273v2” in part of his prototype (though perhaps we’re getting confused about the “ncbi” and “ucsc” versions of the project and which one is the “final” one), and that causes us to worry about using “.”.
If not, I’d like to make a general plea to keep things as compatible as possible with NCBI. I’ve seen a lot of people wasting effort on other tools because of subtle, often unnecessary, naming differences, that make data produced using one version of a reference incompatible with another, essentially identical-except-for-names version of the same reference. Thus, it would nice if a VCF file computed using a reference genome from NCBI would work, w/o “chromosome name remapping” by the user, directly against UCSC.
Perhaps this issue is already addressed by the *.ncbiToUcsc.lift, *.ucsc.to.ncbi.fake.names and *ucscToNcbi.lift files as part of an alias system? Should we be including similar files in our hub/genome?
With NCBI phasing out gi numbers, but not having yet removed them from the .fa and genomes .fna files they produce, I realize that 100% compatibility can be an elusive target to hit. It is nice that the Assembly project .fna’s that Hiram is working from already have the new fasta header format!
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000845245.1_ViralProj14559
>NC_006273.2 Human herpesvirus 5 strain Merlin, complete genome
>gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
>gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
Thanks again to Hiram for sharing his work. That’s a great project that will be really useful to a lot of people!
Brian Lee
Thank you for using the UCSC Genome Browser and building assembly hubs.
It would be wise to generally attempt to use a naming system that restricts to the characters a-zA-Z0-9_ as periods "." and other characters like ":" will cause issues with the underlying MySQL code that is loading databases. Part of the reason for these restrictions is that SQL doesn't allow tables with periods in them because that is how one designates databases with text like "dbName.tableName". There are some existing tickets in our internal system created to attempt to capture these issues and attempt fixes (such as dynamically renaming remote files loaded with name.in.it to use underscores like name_in_it), but please know these differences you have discovered, such as the pipes, are not an attempt by us to restrict naming conventions, rather arriving from underlying software schemas and conventions. We have a new ticket specifically to look into adding an alias file as you mentioned in your previous email.
For the question about placing new Assembly Hubs under the group "Viruses", there was a conscious decision after some consideration to have new assemblies display on the bottom location in attempts to limit confusion between native assemblies and remote assemblies. The decision to to name your group Viruses-HH5 may be the best decision.
Also, please know when you are editing Track Hubs, you can change the time of delay between edits showing up adding the parameter &udcTimeout=10 to your URL where the hub is loaded, this will lower it from the automatic 500 second delay to 10 seconds, so that if you edit the name of a track or hub, it will update in 10 seconds on our site. Later, when you are done developing your hub, and it is no longer changing, it is best to clear this setting back to the default to increase browsing speed. Also, you may be interested to learn how you can add Blat to your assembly hub, and how you can use virtual box software to have local versions of your hub that would not even require internet access using our Genome Browser In a Box (GBiB): http://genome.ucsc.edu/goldenPath/help/hubQuickStartAssembly.html#blat
Thank you again for your inquiry and using the UCSC Genome Browser. If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
All the best,
Brian Lee
UCSC Genomics Institute
--
Hiram Clawson
We are considering alternatives of how to use alternate names for sequences.
The primary difficulty with the names is the variety of context in which they
have to function properly. Yes, work could be done to escape special characters in
places where they need to be escaped, this would be significant work. Here are
some of the places names have to work properly:
1. unix path names (rules out characters such as '/' and others)
2. html text (would not like '&' here, '<' '>' etc...)
3. html URL encodings (no spaces, '?', '%' etc...)
4. MySQL table names (no '.' allowed)
5. MySQL query strings ('_' happens to be a wild card search character,
works in practice, but poses hidden problems)
and probably other environments I'm not thinking about this moment.
After all is said and done, there isn't much left except A-Z a-z 0-9
We are working on it, thanks for your input.
--Hiram
On 4/8/16 1:33 PM, Curtis Hendrickson (Campus) wrote:
> Matthew & Hiram,
>
> A second question occurred to us after the problem with the pipes (|) in the chromosome names: Is there a problem with using period (.) in chromosome names that we have yet to trip over ?
> In our initial testing with chrom="NC_006273.2", things seem to work well.
>
> We noticed that Hiram has converted RefSeq/NCBI's "NC_006273.2" to "NC_006273v2" in part of his prototype (though perhaps we're getting confused about the "ncbi" and "ucsc" versions of the project and which one is the "final" one), and that causes us to worry about using ".".
>
> If not, I'd like to make a general plea to keep things as compatible as possible with NCBI. I've seen a lot of people wasting effort on other tools because of subtle, often unnecessary, naming differences, that make data produced using one version of a reference incompatible with another, essentially identical-except-for-names version of the same reference. Thus, it would nice if a VCF file computed using a reference genome from NCBI would work, w/o "chromosome name remapping" by the user, directly against UCSC.
>
>
> With NCBI phasing out gi numbers, but not having yet removed them from the .fa and genomes .fna<ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses/Human_herpesvirus_5_uid14559/NC_006273.fna> files they produce, I realize that 100% compatibility can be an elusive target to hit. It is nice that the Assembly project .fna's that Hiram is working from already have the new fasta header format!
> ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000845245.1_ViralProj14559
>> NC_006273.2 Human herpesvirus 5 strain Merlin, complete genome
> http://genome-preview.cse.ucsc.edu/gbdb/hubs/refseq/viral/02/GCF_000845245.1_ViralProj14559/GCF_000845245.1_ViralProj14559.ucsc.chrom.sizes
> NC_006273v2 235646
> ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses/Human_herpesvirus_5_uid14559/NC_006273.fna
>> gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
> http://www.ncbi.nlm.nih.gov/nuccore/155573622?report=fasta
>> gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
>
> Thanks again to Hiram for sharing his work. That's a great project that will be really useful to a lot of people!
>
>
> Regards,
> Curtis
>
> Research Associate, Informatics Unit
> Center for Clinical and Translational Science
> http://bioinformatics.uab.edu
> University of Alabama at Birmingham
> Tel: 205.975.5240
>
>
> From: Blair Delane Heater [mailto:bhe...@uab.edu]
> Sent: Friday, April 08, 2016 3:09 PM
> To: Matthew Speir; Curtis Hendrickson (Campus); gen...@soe.ucsc.edu
> Cc: Elliot J Lefkowitz
> Subject: RE: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
>
> Dear Matthew Speir,
>
> Thanks for your help resolving the issue of the unsupported pipes. We attempted to configure our hh5Merlin2 track hub to show up under the Viruses option in the group pull down menu on the Genome Browser Gateway. If we changed the shortLabel in hub.txt to any text other than 'Viruses', it worked and appeared in the drop down menu. But, when the shortLabel was set to 'Viruses', disconnecting and reconnecting the track hub didn't give error but reverted to the shortLabel of previously connected hub. For the moment, we set the shortLabel to be Viruses-HH5(HCMV) as seen below to associate the track hub with Viruses, while we can't successfully place in within that option. Is it possible to set up the track hub under the 'Viruses' option in the group pull down menu? If so, what steps are we missing? If not, is there any standard for group names?
>
>
> Thank you,
> Blair Heater
> From: Matthew Speir<mailto:msp...@soe.ucsc.edu>
> Sent: Wednesday, April 6, 2016 4:22 PM
> To: Curtis Hendrickson (Campus)<mailto:cur...@uab.edu>; gen...@soe.ucsc.edu<mailto:gen...@soe.ucsc.edu>
> Cc: Blair Delane Heater<mailto:bhe...@uab.edu>; Elliot J Lefkowitz<mailto:Ell...@uab.edu>
> Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
>
> Hi Curtis,
>
> Thank you for your question about including pipes in chromosome names in the UCSC Genome Browser.
>
> As you have so far deduced, these pipes, or "|", in the chromosome names are not allowed. In the Genome Browser, your chromosome names should be all alpha or numeric characters, with the first character being a letter.
>
> You suggestion of an alias file is excellent and I have made a note of it in our internal tracking system, however, I can't give an estimate of when this might be implemented.
>
> Matthew Speir
> UCSC Genome Bioinformatics Group
>
> On 4/6/16 10:46 AM, Curtis Hendrickson (Campus) wrote:
> Dear UCSC,
>
> We are working on creating a track hub for the virus Human Herpesvirus 5, based initially on the NCBI RefSeq entry NC_006273.2.
> ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses/Human_herpesvirus_5_uid14559/
>
> this organism has a single segment genome (1 chromosome).
> However, the fasta file provided by NCBI RefSeq, gives a "chromosome name" of
>
> >gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin, complete genome
> For compatibility with RefSeq, we created our hub .2bit file and .bigbed files using that same "chromosome" name, so that VCF files created by users who download their reference genome from RefSeq will work.
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_hub/hub.txt
> This hub passes muster with hubCheck (see below).
>
> However, that seems to be causing some indigestion in the UCSC Browser.
> I can get the display to come up, including the transcript track we provide, but every time we try and interact, it hangs.
> Warning/Error(s):
> OK
>
> And attempts to zoom, show an chromosome name that is truncated after the "gi" - ie at the first pipe (see figure below).
> Questions
>
> 1. Is our guess correct that the pipes (|) in the chromosome name are at fault?
>
> 2. Do you support some sort of "aliases" file like IGV does? (We use that heavily to allow us to use "nicer" chromosome names, while still being compatible with things computed with RefSeq acquired reference fastas)
>
>
> Thanks!
>
> Regards,
> Curtis
>
> Research Associate, Informatics Unit
> Center for Clinical and Translational Science
> http://bioinformatics.uab.edu
> University of Alabama at Birmingham
> Tel: 205.975.5240
>
> Figure: zoom in to UL73 (right click on UL73 transcript, select "zoom to...")
> Browser hangs trying to zoom....
> [cid:image0...@01D18FFE.1D4FF6E0]
>
> --
>
>
Curtis Hendrickson (Campus)
Brian
Thanks for the detailed help.
I can see why things that become paths/table/column names have very restricted character sets. I’m a little surprised that chromosome name in particular becomes a path/table/column name. I would have expected it to be a data value in a column/row, and thus exempt from these restrictions, but I’ve never studies your db schema for tracks/genomes.
Perhaps Hub documentation could be augmented to warn about these restrictions and recommend a standard way of mapping NCBI fasta names, so that most hubs will operate similarly.
I see why you decided to keep the existing groups “closed”. Again, a little more in the docs indicating which field will become that user-visible group name, and noting the restriction, would be great.
The shorter refresh rate will be useful too, and thanks also for the pointers to BLAT and GBiB. We might do BLAT, but not GBiB. .
Regards,
Curtis
Curtis Hendrickson (Campus)
Thanks for the deep explanation. I had not originally realized that *chromosome* names would map directly to database table/column names, rather than mapping down into data values in rows/columns.
Clearly, that architecture imposes a very large number of apparently arbitrary constraints, but I'm sure that architecture decision is not up for discussion at this point in history :-)
Sorry that I have never studied your track schema.
So, yes, I think having alias files like IGV that can be provided as part of a genome (and maybe even as part of a custom tracks), would really help work around the limitation.
Here's the page where it's described: https://www.broadinstitute.org/igv/LoadData (at bottom). They also implement some automatic regex based re-mappings, too, to cover common cases:
"Note: Certain well-known aliases are built into IGV and do not require an alias file. These include mappings that involve adding or removing the prefix "chr" to the name, for example 1 -> chr1 and chr1 -> 1. Also, NCBI identifiers that start with "gi|" and follow the pattern illustrated in the example above are automatically mapped."
We will change to NC_#####v2.
However, I'm less clear on why the "_" would cause match problem. In MySQL, that only happens in the context of LIKE but not in the context of =, so it's not clear why that would be such an issue.
Thanks again for all your detailed and rapid help!
>> gi|155573622|ref|NC_006273.2| Human herpesvirus 5 strain Merlin,
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.tx
> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt/bitmap does not already exist, creating.
> reading http/https/ftp data - 406 bytes at 0 - on
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.tx
> t hub
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
> shortLabel hh5Merlin2
> longLabel Track hub for HCMV Strain Merlin based on
> gi|155573622|ref|NC_006273.2| genomes.txt has 1 elements
> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pu
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2
> hubCheck/https/data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/trackDb.txt/bitmap does not already exist, creating.
> reading http/https/ftp data - 143 bytes at 0 - on
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2
> /trackDb.txt checking _hh5Merlin2._hh5Merlin2_refseq_mrna type bigBed
> 12 at
> /NC_006273.2.mrna.bb
> udcfileOpen(https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pu
> on
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2
Hiram Clawson
Also remember, there is the complicating factor of actual humans wanting
to use these names. Would you rather think about a sequence named CM000663.2
or the UCSC name of 'chr1', or 'gi|568336023|gb|CM000663.2|' ? Again, you
are correct, it is a translation issue which should
be taken care of by software, but the translation has to take place somewhere.
The practice to date has been that UCSC establishes the names of sequences,
UCSC provides the download files, users pick up UCSC files and run analysis pipelines
on UCSC data to display in the UCSC genome browser. Yes, NCBI/Genbank/INSDC
is the ultimate source of the data to begin with, but the names there are
not the most convenient for a variety of purposes.
The practice is evolving to continue to use UCSC names in established genomes
already on the genome browser, but newer genome assemblies will most likely
use a combination of names supplied from NCBI/Genbank/INSDC. At this time
my focus is on the names included in the _assembly_report.txt file with
the NCBI assembly download files, and the chr2acc name translation files
in the Primary_Assembly directory hierarchy. This can lead to UCSC 'chrN'
'chrUn_*' 'chr*_random' types of names for assembled chromosome types of
assemblies. However, many
assemblies these days are merely piles of unplaced scaffolds, and the naming
scheme there is to merely replace the '.' with a 'v' in the accession name when
the assembly is going to be in a UCSC MySQL database. I have experimented with leaving
the accession name with the '.' when the assembly is only in an assembly hub.
So far, this appears to work well enough.
You are correct, naming issues are a headache. I suspect they will continue thusly.
--Hiram
Curtis Hendrickson (Campus)
Matthew, Hiram,
We're trying to add a BLAT server to our viral custom track hub.
We set up the server on a publically visible VM and it seems to be running fine (see ps on VM below).
But when I try to query it, I get errors, both from the command-line and the website, but I can’t figureout how I’ve mis-configured it.
It looks like it’s slapping the URL of the seq_dir infront of the local path of the seq_dir, but I’m not sure where that’s coming from!
All help appreciated!
Regards,
Curtis
Research Associate, Informatics Unit
Center for Clinical and Translational Science
University of Alabama at Birmingham
Tel: 205.975.5240
Email: cur...@uab.edu
Regards,
Curtis
Hub: https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hub.txt
##
## UCSC blat error
##
Warning/Error(s):
##
## gfClient error
##
# telnet connection ok
curtish@genome-BMIlinux:~/vm-blat4trackhub-blat$ telnet 164.111.161.69 17777
Trying 164.111.161.69...
Connected to 164.111.161.69.
Escape character is '^]'.
^CConnection closed by foreign host.
# blat errors out
curtish@genome-BMIlinux:~/vm-blat4trackhub-blat$ ucsc_kent/2016-05-10/blat/gfClient 164.111.161.69 17777 data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2 test.fa test.psl
Expecting 6 words from server got 2
##
## gfServer running on VM, can see .2bit files
##
# process running with recommended flags
blat@blat4trackhubs6-open:~$ ps -eaf | grep gfServer
blat 23003 1 0 21:29 pts/1 00:00:00 ucsc_kent/2016-05-10/blat/gfServer start localhost 17777 -trans data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
blat 23004 1 0 21:29 pts/1 00:00:00 ucsc_kent/2016-05-10/blat/gfServer start localhost 17779 -stepSize=5 data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
blat 23827 21461 0 21:33 pts/1 00:00:00 grep --color=auto gfServer
# 2bit files fine
blat@blat4trackhubs6-open:~$ file data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit: data
data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit: data
blat@blat4trackhubs6-open:~$


Hiram Clawson
Go to the directory where the 2bit file exists. Use that as your
current working directory. Run gfServer on the file name KP745636v1.2bit
without any extra directory path. See also:
https://genome.ucsc.edu/goldenpath/help/hubQuickStartAssembly.html#blat
--Hiram
On 5/25/16 2:47 PM, Curtis Hendrickson (Campus) wrote:
> Matthew, Hiram,
>
>
>
> We're trying to add a BLAT server to our viral custom track hub.
>
>
>
> We set up the server on a publically visible VM and it seems to be running fine (see ps on VM below).
>
> But when I try to query it, I get errors, both from the command-line and the website, but I can't figureout how I've mis-configured it.
>
> It looks like it's slapping the URL of the seq_dir infront of the local path of the seq_dir, but I'm not sure where that's coming from!
>
>
>
> All help appreciated!
>
> Regards,
> Curtis
>
> Research Associate, Informatics Unit
> Center for Clinical and Translational Science
> http://bioinformatics.uab.edu
> University of Alabama at Birmingham
> Tel: 205.975.5240
> blat 23004 1 0 21:29 pts/1 00:00:00 ucsc_kent/2016-05-10/blat/gfServer start localhost 17779 -stepSize=5 data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
>
> blat 23827 21461 0 21:33 pts/1 00:00:00 grep --color=auto gfServer
>
>
>
> # 2bit files fine
>
> blat@blat4trackhubs6-open:~$ file data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
>
> data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit: data
>
> data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit: data
>
> blat@blat4trackhubs6-open:~$
>
>
>
>
> [cid:image0...@01D1B6A5.2CCC4A60]
Hiram Clawson
Are all your blat servers running on the same port numbers ?
They need to be on different port numbers. This is how the gfClient
can identify the blat server, by the port number. Each entry for
each genome blat server will be unique since they must all have
different port numbers.
Your subdirectory for each genome is a correct way to do that.
It is OK to symlink them all together for the gfServer operations,
just start each gfServer on a different port.
The directory paths and locations of 2bit files and track definition
files are arbitrary. They could all be together in one directory, but that
would be messy. It is more orderly to separate genomes out by
directory path.
Keep me advised of what works or not.
--Hiram
On 5/26/16 8:06 AM, Curtis Hendrickson (Campus) wrote:
> Hiram,
>
>
>
> Thanks. The problem is that our hub has multiple genomes, each in it's own subdirectory.
>
> Essentially, for each genome we have a subdirectory with the 2bit and all the track files.
>
>
>
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit
>
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
>
>
>
> I symlink'ed them all into one location on the BLAT VM, but I get an odd error:
>
>
>
> I blat against one genome and get an error for the other
>
> [cid:image0...@01D1B735.BBFB3FD0]
>
> Warning/Error(s):
>
> Couldn't open https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/KP745636v1.2bit
>
>
>
>
>
> We have the correct paths in the genomes.txt (see below). Does the system really require that only the tracks be in per-genome subdirectories, and all .2bits must be at the top level?
>
>
>
> It seems like we've violated some hub layout assumption we don't quite understand.
>
>
>
> Also, do we need to repeate the Blat lines for each genome? Is there a way to specify this at the hub level?
>
>
>
> Thank you,
>
> Curtis
>
>
>
> https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/genomes.txt
> genome hh5Merlin2
> trackDb hh5Merlin2/trackDb.txt
> twoBitPath hh5Merlin2/NC_006273v2.2bit
> groups hh5Merlin2/groups.txt
> description NC_006273v2
> organism HH5 strain Merlin
> defaultPos NC_006273v2:1-235646
> orderKey 100
> scientificName Human herpesvirus 5
> htmlPath https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/description.html
> transBlat 164.111.161.69 17777
> blat 164.111.161.69 17779
>
> genome hh5BE_7_2011v1
> trackDb hh5BE_7_2011v1/trackDb.txt
> twoBitPath hh5BE_7_2011v1/KP745636v1.2bit
> groups hh5BE_7_2011v1/groups.txt
> description KP745636v1
> organism HH5 strain BE/7/2011
> defaultPos KP745636v1:1-237117
> orderKey 100
> scientificName Human herpesvirus 5
> htmlPath https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/description.html
> transBlat 164.111.161.69 17777
> blat 164.111.161.69 17779
>
>
>
>
>
>
> -----Original Message-----
> From: Hiram Clawson [mailto:hi...@soe.ucsc.edu]
> Sent: Wednesday, May 25, 2016 6:10 PM
> To: Curtis Hendrickson (Campus); Blair Delane Heater; Matthew Speir; gen...@soe.ucsc.edu
> Cc: Elliot J Lefkowitz
> Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
>
>
>
>
>
>
> Go to the directory where the 2bit file exists. Use that as your current working directory. Run gfServer on the file name KP745636v1.2bit without any extra directory path. See also:
>
> https://genome.ucsc.edu/goldenpath/help/hubQuickStartAssembly.html#blat
>
>
>
> --Hiram
>
>
>
> On 5/25/16 2:47 PM, Curtis Hendrickson (Campus) wrote:
>
>> Matthew, Hiram,
>
>>
>
>>
>
>>
>
>> We're trying to add a BLAT server to our viral custom track hub.
>
>>
>
>>
>
>>
>
>> We set up the server on a publically visible VM and it seems to be running fine (see ps on VM below).
>
>>
>
>> But when I try to query it, I get errors, both from the command-line and the website, but I can't figureout how I've mis-configured it.
>
>>
>
>> It looks like it's slapping the URL of the seq_dir infront of the local path of the seq_dir, but I'm not sure where that's coming from!
>
>>
>
>>
>
>>
>
>> All help appreciated!
>
>>
>
>> Regards,
>
>> Curtis
>
>>
>
>> Research Associate, Informatics Unit
>
>> Center for Clinical and Translational Science
>
>> http://bioinformatics.uab.edu University of Alabama at Birmingham
>
>> Tel: 205.975.5240
>
Curtis Hendrickson (Campus)
Hiram,
Thanks. The problem is that our hub has multiple genomes, each in it's own subdirectory.
Essentially, for each genome we have a subdirectory with the 2bit and all the track files.
https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP745636v1.2bit
https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_006273v2.2bit
I symlink'ed them all into one location on the BLAT VM, but I get an odd error:
I blat against one genome and get an error for the other

Warning/Error(s):
Couldn't open https://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/KP745636v1.2bit
-----Original Message-----
From: Hiram Clawson [mailto:hi...@soe.ucsc.edu]
Sent: Wednesday, May 25, 2016 6:10 PM
To: Curtis Hendrickson (Campus); Blair Delane Heater; Matthew Speir; gen...@soe.ucsc.edu
Cc: Elliot J Lefkowitz
Subject: Re: [genome] adding a genome using a custom track hub - are pipe's allowed in chr names?
Good Afternoon Curtis:
> data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP7
> 45636v1.2bit
> data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5Merlin2/NC_0062
> 73v2.2bit
>
> data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/hh5BE_7_2011v1/KP7
> 45636v1.2bit: data
Curtis Hendrickson (Campus)
Thanks for the speedy reply.
This implies that one must run one pair of blat server processes per genome, and thus open one pair of ports per genome.
Since we're planning to serve hundreds of genomes, that seems like a *huge* IT headache and overhead.
As gfServer seems to be able to take a list of .2bit files (genomes? Contigs?) we were hoping to have one server for the hub.
Since our genomes are all different strains of the same species, being able to BLAT across all genomes is a good thing, but maybe things aren't/shouldn't be set up to work that way?
How do you solve this for your Genome Assemblies project?
It seems like the BLAT/ gfClient at your end is not using the paths in the genomes.txt file....
How does that process construct the path it sends to gfServer?
Also, for debugging, is there a way to get the gfServer at my end to show me the request coming in ? Despite these errors, my gfServer log files remain empty.
Regards,
Curtis
Hiram Clawson
Correct, one gfServer blat process for one genome. This is a bottleneck, no doubt.
At this time we have no solution to this situation. gfServer/gfClient would need to rewritten
to allow multiple genomes in one instance. We have been discussing such an idea here
for some time.
gfClient/browser requests know nothing about paths to any file. All they know is a port
number which they use to communicate with the gfServer. When the browser needs sequence
for the genome for display purposes, then it uses the path names from the hub definition
files. Please note, the references to files from the hub definition files are relative
references from the definition file itself. They do not need to be full path names.
That would make it difficult to copy the hub to a new location.
For example, in your hub.txt file, the reference:
descriptionUrl file://data.genome.uab.edu/public/ucsc_track_hubs/hcmv_pub/description.html
Should simply be:
descriptionUrl description.html
Same for your htmlPath in your genomes.txt file.
There are several options to gfServer to log everything in detail, for example:
-log=logFile Keep a log file that records server requests.
-seqLog Include sequences in log file (not logged with -syslog).
-ipLog Include user's IP in log file (not logged with -syslog).
-syslog Log to syslog.
--Hiram
Curtis Hendrickson (Campus)
With one genome per blat server pair, everything works!
Thanks for the clarifications. You don't always give me the answers I hope for, but you do give me the correct answers, which is invaluable :-)
That one-server-per-assembly is going to be a big burden. I have to open IT tickets to get ports opened to the outside world, and I'll need a much bigger VM to run 1,000 gfServer processes, even if they're all idle 99.9% of the time.
It sounds like a protocol where a relative path or genome name argument could be sent to the gfServer that would limit queries to the desired relative path might solve the problem. Yes, it's always hard to change line-protocols for client-server apps!
So, in my case, the gfServer returned results for both of the .2bits I had loaded (one for each genome), then the UCSC server runs off looking for those .2bit files in our hub, and finds only the one for the genome it had thought it requested. So, it's the results from the 2nd genome that break everything. And even if it found the .2bit file, it would be like getting combined hits from hg18 and hg19 at the same time - not something the system is set up to handle correctly.
Also, thanks for pointing out the absolute path in descriptionUrl & htmlPath - I get the relative path thing; they have been fixed.
Finally, of course, two more questions (do you want me to start a new thread/subject line for each question, or is this easier?)
** [In-Silico PCR] **
I did Tools>In Silico PCR, and got
"Note: In-Silico PCR is not available for hub_86647_HH5 strain Merlin NC_006273v2; defaulting to Human Dec. 2013 (GRCh38/hg38)"
Is that easy to add? What is required? It looked from hubQuickStartAssembly that setting up the gfServers would enable both blat and In-Silico PCR.
** hh5Merlin2.chrom.sizes **
I notice Blair put the .chrom.sizes files at the top of the hub, not in the assembly/genome specific subdirectories.
Can they be moved into the subdirectories?
Cath Tyner
* Unsubscribe: Email genome-announ...@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser discussion list" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
Cath Tyner
In-Silico PCR
I did Tools>In Silico PCR, and got
"Note: In-Silico PCR is not available for hub_86647_HH5 strain Merlin NC_006273v2; defaulting to Human Dec. 2013 (GRCh38/hg38)"
Is that easy to add? What is required? It looked from hubQuickStartAssembly that setting up the gfServers would enable both blat and In-Silico PCR.
hh5Merlin2.chrom.sizes
I notice Blair put the .chrom.sizes files at the top of the hub, not in the assembly/genome specific subdirectories.
Can they be moved into the subdirectories?
* Unsubscribe: Email genome-announ...@soe.ucsc.edu
Cath
Actually, no, we’ve stayed “on list”, and I haven’t heard from Hiram since I sent the inquiry you forward below.
(You had mentioned he was on vacation).
I can re-send later today….
Curtis
Curtis Hendrickson (Campus)
Cath Tyner
In-Silico PCR
I did Tools>In Silico PCR, and got
"Note: In-Silico PCR is not available for hub_86647_HH5 strain Merlin NC_006273v2; defaulting to Human Dec. 2013 (GRCh38/hg38)"
Is that easy to add? What is required? It looked from hubQuickStartAssembly that setting up the gfServers would enable both blat and In-Silico PCR.
hh5Merlin2.chrom.sizes
I notice Blair put the .chrom.sizes files at the top of the hub, not in the assembly/genome specific subdirectories.
Can they be moved into the subdirectories?
* Unsubscribe: Email genome-announ...@soe.ucsc.edu
