dbSNP variant missing from UCSC dbSNP 147 track
Tam, Oliver
Dear Genome Browser team,
I was looking for a variant at the splice donor of PMS2 (chr7:6017218, G>T [cDNA], C>A [genomic]), and noticed on the UCSC genome browser that, while there was a entry in the ClinVar track, there is no corresponding entry in the dbSNP (147) track (see screenshot 1). I then clicked the ClinVar entry (screenshot 2), and found the dbSNP entry (876661113), which I could click to access it. I was just surprised to not see it in the all SNPs (147) and Flagged SNPs (147) tracks, and was wondering if it is not displaying properly on my end.
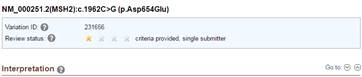
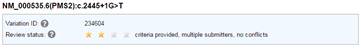
I also tried to access the ClinVar variation report from screenshot 2. It redirected me to this URL (http://www.ncbi.nlm.nih.gov/clinvar/variation/231656/), which is a different variant (screenshot 3). However, if I click on the ClinVar Allele submission links, they link to the correct submission, which then allowed me to go to the correct ClinVar page (http://www.ncbi.nlm.nih.gov/clinvar/variation/234604) (screenshot 4). I’m wondering if there’s something on my end that’s causing this error.
Please let me know if there are any questions or confusion, or if you cannot replicate my issue.
Thanks for all your help.
Cheers,
Oliver
Screenshot 1
![]()

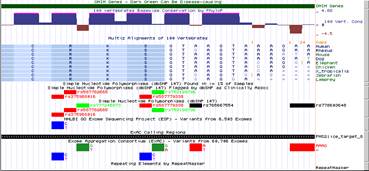
Screenshot 2

Screenshot 3
Screenshot 4
-------------------------------------------------------------------
The information transmitted by this e-mail and any included
attachments are from ARUP Laboratories and are intended only for the
recipient. The information contained in this message is confidential
and may constitute inside or non-public information under
international, federal, or state securities laws, or protected health
information and is intended only for the use of the recipient.
Unauthorized forwarding, printing, copying, distributing, or use of
such information is strictly prohibited and may be unlawful. If you
are not the intended recipient, please promptly delete this e-mail
and notify the sender of the delivery error or you may call ARUP
Laboratories Compliance Hot Line in Salt Lake City, Utah USA at (+1
(800) 522-2787 ext. 2100
Tam, Oliver
Dear Genome Browser team,
I forgot to clarify that I’m using the hg19/GRCh37 build of the human genome.
Thanks.
Cheers,
Oliver
Luvina Guruvadoo
If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
--
