modify some Webpages of UCSC local mirror
15 views
Skip to first unread message
Fu weiwei
Sep 28, 2017, 11:24:51 AM9/28/17
to gen...@soe.ucsc.edu
Dear UCSC Genome Browser Team,
I have built an UCSC local mirror but something about the code and configuration file is bothering me. I will appreciate if you can help me to deal with these following problems.
(1) How to add a simple button on the hgTracks page that can export images with PDF/PS format.
I know the navigation bar (View) of UCSC mirror page have this function. But now I have a new navigation bar of my web. So I want to only add a button with the same function of View (PDF/PS) that originally exists on the navigation bar of hgTracks page. Like this (see figure below) but not on the navigation bar. In addition, I also want to know how to remove the grid lines by setting this page.
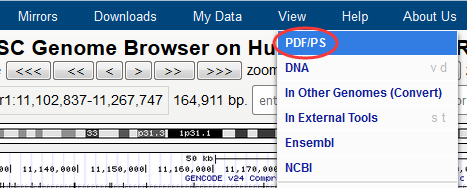
(2) How to modify the background color of the hgTables page.
I want to introduce the hgTables page into my web. However, the background color of this page is very different from my website. I guess it may be written in the HGStyle.css file. But I can't modify the bgcolor although I did a lot of attempts to rewrite this CSS file. Can you help me point out the definition position of bgcolor?

Waiting for your reply! Thank you very much for your time.
All the best,
Weiwei Fu
发自网易邮箱大师
Cath Tyner
Oct 4, 2017, 4:48:54 PM10/4/17
to Fu weiwei, gen...@soe.ucsc.edu
Hello Weiwei,
Thank you for contacting the UCSC Genome Browser support team. Below are answers to your questions:
1. PDF/PS button
Adding a button involves changing the C code. Look in kent/src/hg/hgTracks/hgTracks.c for a comment "/* Make line that says position. */". About 25 lines below that (currently line 8123) is a line that says "hButton("hgt.jump", "go");". Add a button or link after that. Code for forming a URL to get PDF/PS output is in hgTracks/menu.c:
char *menuStr, buf[4096], uiVars[1024];safef(uiVars, sizeof(uiVars), "%s=%s", cartSessionVarName(), cartSessionId(cart));...safef(buf, sizeof(buf), "../cgi-bin/hgTracks?%s&hgt.psOutput=on", uiVars);
2. Hide grid lines
From the browser (http://genome.ucsc.edu/cgi-bin/hgTracks),
- Click "Genome Browser" at the top blue horizontal menu > "Configure."
- Under "Configure Image", uncheck "Show light blue vertical guidelines, or light red vertical window separators in multi-region view"
- Press submit.
3. Changing Table Browser colors
Change colors in the following two files:
- ~/kent/src/hg/htdocs/style/HGStyle.css
- ~/kent/src/hg/inc/hgColors.h
In HGStyle.css, make changes in the following two places:
a.
body.cgi {margin: 0;padding: 0;- background: #FFF9D2;+ background: #FFFFFF;}
b.
/* A subheading bar is the light blue bar that appears at the top of web.c section boxes */-.subheadingBar {font-weight: bold; font-size:1.1em; color:#000000; background-color:#D9E4F8;+.subheadingBar {font-weight: bold; font-size:1.1em; color:#000000; background-color:#FFFEE8;
Save the files and do the 'make' in src/hg/htdocs/style
Then, change colors in the lines below, in hgColors.h:
Be aware that the changes in this file will also affect other tools, such as the Variant Annotation Integrator.
-#define HG_COL_OUTSIDE "FFF9D2"-#define HG_CL_OUTSIDE 0xFFF9D2-#define HG_COL_INSIDE "FFFEE8"-#define HG_CL_INSIDE 0xFFFEE8+#define HG_COL_OUTSIDE "FF0000"+#define HG_CL_OUTSIDE 0xFF0000+#define HG_COL_INSIDE "FFFFFF"+#define HG_CL_INSIDE 0xFFFFF
Then, do the top level 'make' in 'hg':
cd ~/kent/src/hg'make clean && make -j4'
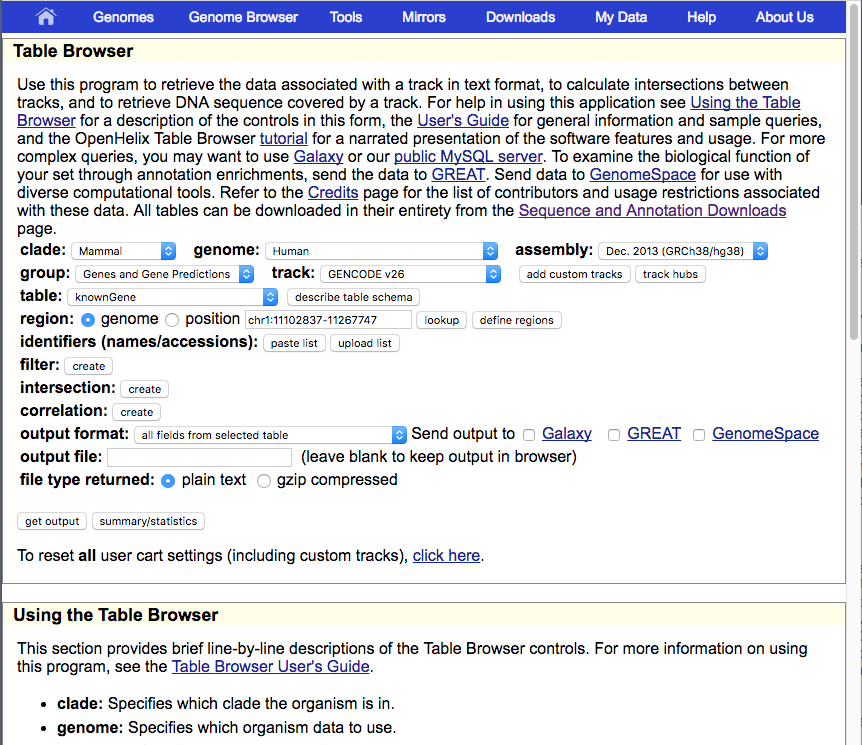
As an example, the changes above will result in a Table Browser that looks like this:
Finally, please be aware that UCSC Genome Browser products are offered are free for non-commercial use by non-profit organizations, academic institutions, and for personal use. Commercial use requires purchase of a license with setup fee and annual payment. A non-profit organization in service to a for-profit organization must purchase a commercial license, even if the latter already possesses a commercial product license. See more information at our UCSC Genome Browser Store.
Please respond to this list if you have further questions!
* Unsubscribe: Email genome-announce+unsubscri...@soe.ucsc.edu
Thank you for contacting the UCSC Genome Browser support team.
Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private help: Email
genom...@soe.ucsc.eduUCSC Genome Browser Announcements List (email alerts for new data & software):
* Subscribe: Email genome-announce+subscribe...@soe.ucsc.edu * Unsubscribe: Email genome-announce+unsubscri...@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/5104cafd.6f8e.15ec8b07956.Coremail.weiweifu666%40126.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
Fu weiwei
Oct 9, 2017, 11:50:47 AM10/9/17
to Cath Tyner, gen...@soe.ucsc.edu
Hi Cath,
Thank you for your reminder. The UCSC local mirror that our team built is just for academy research and non-commercial.
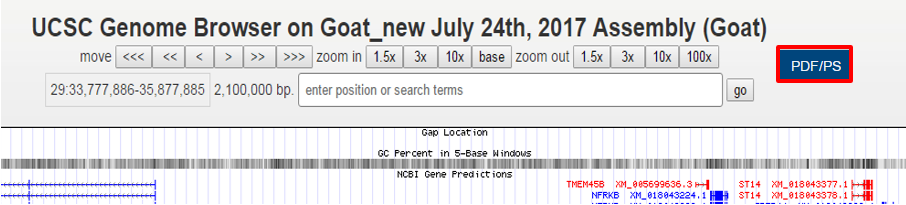
I am very grateful for your help and reply. However, I can't solve the first question because I only mastered programming languages including html, Javascript, php and mysql but not including the C. So I didn't know how to put the code (for forming a URL to get PDF/PS output in hgTracks/menu.c) into hgTracks.c. Now I can only add a button (see figure below) on the hgTracks page but this button can't export images. I will be very grateful if you can supply the altered detailed code or give me a compiled binary file (hgTracks).

Thank you for your replication. Any help will be helpful for me.
All the best,
Weiwei Fu
发自网易邮箱大师
Cath Tyner
Oct 9, 2017, 4:47:55 PM10/9/17
to Fu weiwei, gen...@soe.ucsc.edu
Hello Weiwei,
Due to limited resources, the UCSC Genome Browser support team generally can't accommodate support requests for programming or scripting help. Sometimes we are able to help with limited scripting assistance; these are usually small scripts to assist with file manipulation. We can continue to provide general direction regarding mirror site customization, but we don't have the resources to provide detailed programming help. In cases such as this, you might be able to find assistance though other help sites such as Stack Overflow or Biostars. Good luck with your customization, and please feel free to respond to this forum with general follow-up questions.
Due to limited resources, the UCSC Genome Browser support team generally can't accommodate support requests for programming or scripting help. Sometimes we are able to help with limited scripting assistance; these are usually small scripts to assist with file manipulation. We can continue to provide general direction regarding mirror site customization, but we don't have the resources to provide detailed programming help. In cases such as this, you might be able to find assistance though other help sites such as Stack Overflow or Biostars. Good luck with your customization, and please feel free to respond to this forum with general follow-up questions.
If you do find a solution, you can reply to this forum so that your answer posts to the archives; this would assist other users in the future who might search for this topic.
Thank you for contacting the UCSC Genome Browser support team.
Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private help: Email
genom...@soe.ucsc.eduUCSC Genome Browser Announcements List (email alerts for new data & software):
* Subscribe: Email genome-announce+subscribe...@soe.ucsc.edu * Unsubscribe: Email genome-announce+unsubscri...@soe.ucsc.edu
Reply all
Reply to author
Forward
0 new messages
