GTF file of Introns
Andy Rampersaud
--
Graduate Student, Bioinformatics
Waxman Lab, Boston University
Jonathan Casper
Hello Andy,
Thank you for your question about generating GTF files of intron regions. There is a way to generate a GTF file that contains the intron regions of a track, but it will be a two-step process. Due to the mechanics of how GTF files work, the result will be a GTF file that picks out the intron regions of your data, but they will be called exons within the GTF file itself.
The basic overview of what we will do here is take your track data and use the Table Browser to create a custom track that just contains the introns from your data. Then we will use the Table Browser a second time to generate GTF output from that custom track.
1. Load your data file as a custom track.
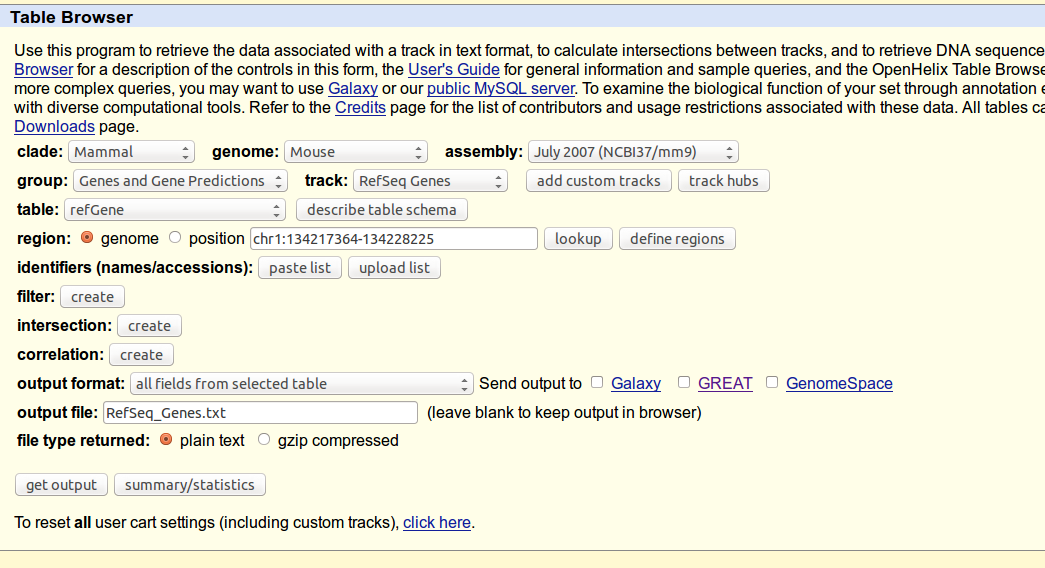
2. Open the UCSC Table Browser at http://genome.ucsc.edu/cgi-bin/hgTracks and select your custom track.
3. Set up any region and identifier filters that you want to impose (or just set the region to "genome"), and set the output format to "custom track".
4. Click "get output".
5. On the next page, name your new custom track (e.g., "introns from my data") and select the box to create one BED record per "Introns plus 0 bases on each end".
6. Click "get custom track in table browser". You should now be returned to the main UCSC Table Browser page.
7. Select your new custom track from the menu (e.g., Group: Custom Tracks, Track: introns from my data).
8. Change the output format to "GTF - gene transfer format".
9. Click "get output".
The result should be a GTF file that describes the intron regions of your data.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu or genome...@soe.ucsc.edu. Questions sent to those addresses will be archived in publicly-accessible forums for the benefit of other users. If your question contains sensitive data, you may send it instead to genom...@soe.ucsc.edu.
--
Jonathan Casper
UCSC Genome Bioinformatics Group
--
Andy Rampersaud
#Get all the unique accession numbers:
#length(unique(Gene_List$"name"))
#[1] 33555
#length(unique(RefSeq_GTF$"gene_id"))
#[1] 33599
#length(unique(RefSeq_GTF$"transcript_id"))
#[1] 34599
Jonathan Casper
Hello Andy,
The GTF output from the UCSC Table Browser is a bit limited, in that it re-uses the transcript ID to fill the gene ID column. You can obtain a GTF file with actual gene identifiers (from the "name2" field of the refGene table) by using the genePredToGtf utility as described at http://genomewiki.ucsc.edu/index.php/Genes_in_gtf_or_gff_format.
Regarding the counts you found for unique identifiers, I have a few comments. I'm not sure how R arrived at the figure of 33555 unique names in the table dump - that does not match what I see in the data. The number should be 33599. The reason that there are an extra 1000 transcript IDs (for a total of 34599) is that GTF transcript identifiers are required to be unique. When we have mapped a transcript to multiple locations in the genome (e.g., NM_001025241), then the Table Browser must generate additional transcript identifiers for the alternate locations in GTF output. Those identifiers take the form of NM_001025241_dup1, NM_001025241_dup2, and so on. The Table Browser does not generate alternate gene names. For contrast, the output of genePredToGtf contains 34675 distinct transcript names - more than the output from the UCSC Table Browser. In this case, it looks like there are some transcripts for which genePredToGtf gave alternate identifiers while the Table Browser (for some reason) did not.
Overall, we recommend using the genePredToGtf output.
--
Jonathan Casper
UCSC Genome Bioinformatics Group
Andy Rampersaud
I just wanted to follow up on the issue of unique accession number counts. I'm confident that R is correctly counting the number of unique accession numbers. I think the issue is the refGene table (when downloaded from the UCSC Table Browser) is missing these 44 accession numbers. I did a comparison between the Table_List of accession numbers and GTF_List of accession numbers:
#length(intersect(Table_List,GTF_List))
#[1] 33555
#length(setdiff(Table_List,GTF_List))
#[1] 0
#length(setdiff(GTF_List,Table_List))
#[1] 44
Jonathan Casper
Hello Andy,
Sorry for the delay on this. The number of entries in the refGene table is steadily changing, as we regularly incorporate new additions to the RefSeq library of transcripts. Right now, we count 24327 distinct gene symbols in the refGene table for the mm9 assembly. Your count of 24197 sounds close enough to be accurate if you downloaded your dataset some time ago (as seems likely given the difference in transcript counts).
Using the refGene table dump as an index to insert gene symbols into your GTF file sounds like it should work just fine; the transcript names in each file are coming from the same data source anyway.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu or genome...@soe.ucsc.edu. Questions sent to those addresses will be archived in publicly-accessible forums for the benefit of other users. If your question contains sensitive data, you may send it instead to genom...@soe.ucsc.edu.
--
Jonathan Casper
UCSC Genome Bioinformatics Group
