Questions about Interpreting Multiz Alignment of 46 Vertebrates
Ngo, Kathie J.
Hi ,
I have a novice question about interpreting the 46ways cons data. From the screenshot below, does the “=” symbol for the Gorilla alignment (see yellow highlight in screenshot) stand for alignment gap?
Please also correct me if my understanding of the following is incorrect:
· Light blue tracks match the reference, “C”
· Blank white tracks (i.e. Wallaby, Stickleback, and Lamprey) are missing in this 46ways alignment
· The N in the Mouse_lemur track could be any nucleotide (A,C,G, or T)
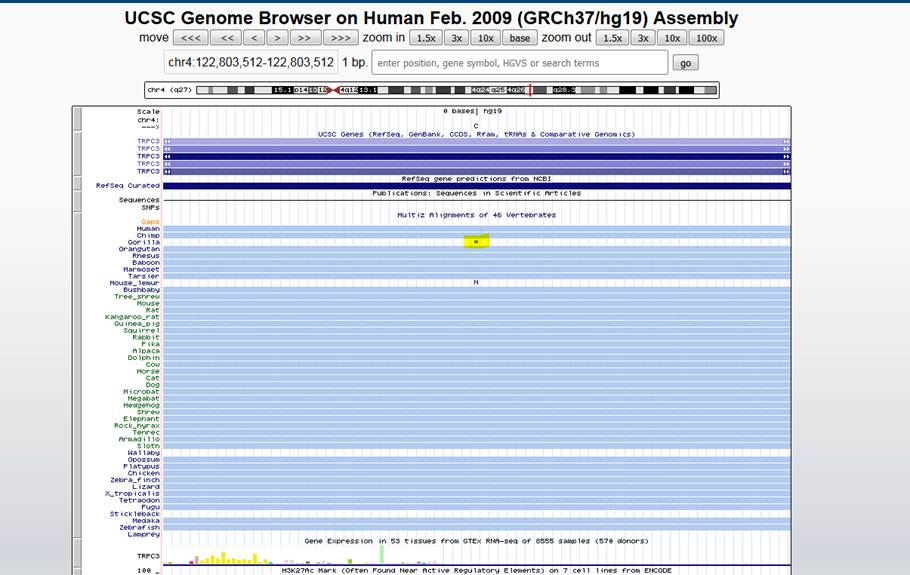
Thank you for your time and help.
Best Regards,
Kathie Ngo
UCLA HEALTH SCIENCES IMPORTANT WARNING: This email (and any attachments) is only intended for the use of the person or entity to which it is addressed, and may contain information that is privileged and confidential. You, the recipient, are obligated to maintain it in a safe, secure and confidential manner. Unauthorized redisclosure or failure to maintain confidentiality may subject you to federal and state penalties. If you are not the intended recipient, please immediately notify us by return email, and delete this message from your computer.
Jairo Navarro Gonzalez
Hello Kathie,
Thank you for using the UCSC Genome Browser and your inquiry.
The information from the track description page for this track contains the answers to your questions.
http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=cons46way
For example, from that page, you can find the following information:
Double line: Aligning species has one or more unalignable bases in the gap region. Possibly due to excessive evolutionary distance between species or independent indels in the region between the aligned blocks in both species.
Vertical blue bar: Represents a discontinuity that persists indefinitely on either side, e.g. a large region of DNA on either side of the bar comes from a different chromosome in the aligned species due to a large scale rearrangement.
If you are new to the UCSC Genome Browser, I would recommend viewing some of our tutorial videos, http://genome.ucsc.edu/training/.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu.
All messages sent to that address are archived on a publicly-accessible Google Groups forum.
If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
Jairo Navarro
UCSC Genomics Institute
Want to share the Browser with colleagues?
Host a workshop: http://bit.ly/ucscTraining
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CY1PR12MB09044AC494F5D2D104297293E1BA0%40CY1PR12MB0904.namprd12.prod.outlook.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
