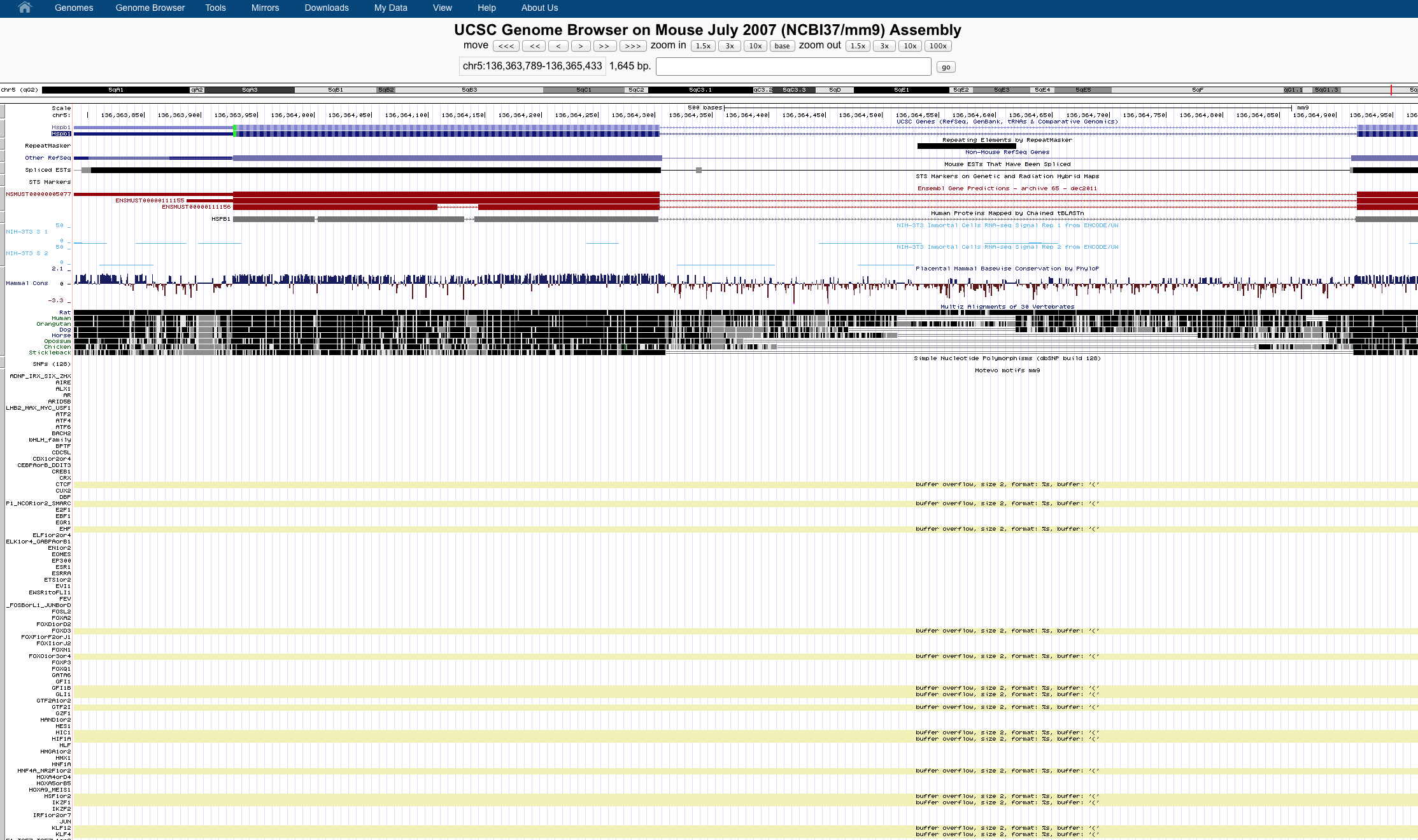
Buffer overflow errors when uploading trackhubs of bigbeds
Jake Yeung
Cath Tyner
Once we have more information, we can take a closer look and hopefully provide suggestions for fixing this issue.
* Unsubscribe: Email genome-announce+unsubscri...@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/03E6462D-60C4-4D98-ABFC-954CF09551CD%40gmail.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
Christopher Lee
Hi Jake,
Thank you for your question about buffer overflows when loading your track hub. It appears that some lines of your bigBed files don't contain a strand column, and thus are not valid bigBed 6 files, which is causing the errors you are seeing.
We also recommend using the hubCheck utility (available here: http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/hubCheck) in order to check the tracks in your hubs.
If you modify your bigBed files so they are valid bed 6 files your hub should work.
Please let us know if you have any further questions!
Thank you again for your inquiry and using the UCSC Genome Browser. If
you have any further questions, please reply to gen...@soe.ucsc.edu.
All messages sent to that address are archived on a
publicly-accessible forum. If your question includes sensitive data,
you may send it instead to genom...@soe.ucsc.edu.
Christopher Lee
UCSC Genomics Institute
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CAC4PgmJ1FHVc9atNu4Su56aogfUcCoHPvJe8RAK%3DQ9J9Gwc12g%40mail.gmail.com.
