Vertebrate Multiz Alignment & Conservation (46 Species)
杨振军(Zhenjun Yang)
Hi,
I am a researcher from BGI, and I want to get the amino acid sequence in Vertebrate Multiz Alignment & Conservation (46 Species) ,but can only get the DNA sequence now,
I wonder if you would tell me how to set the track to get the amino acid sequence. I am looking forward to hearing from you. Thank you very much !
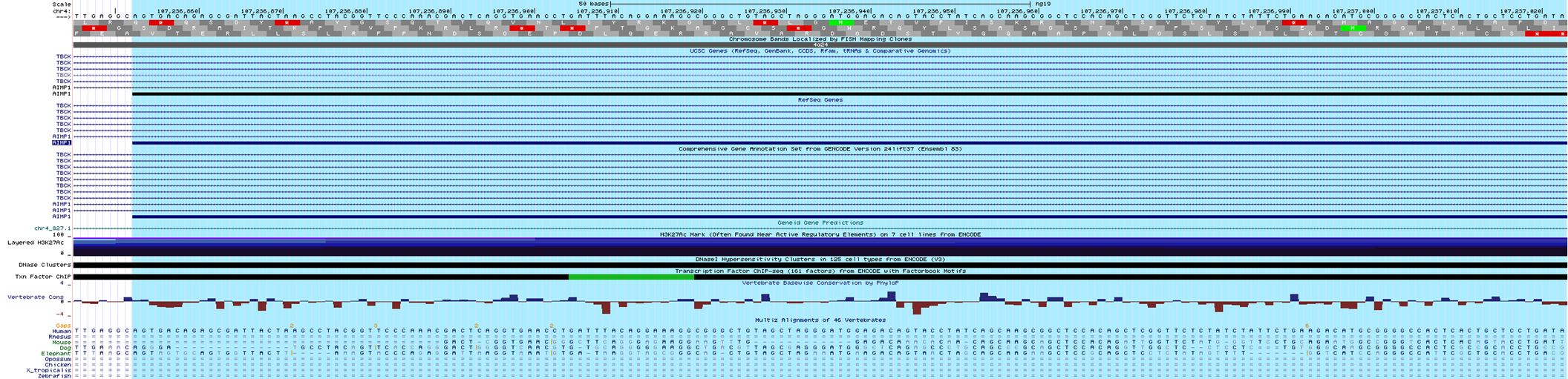
Best,
Sincerely,
Zhenjun Yang
Cath Tyner
* Unsubscribe: Email genome-announce+unsubscri...@soe.ucsc.edu
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/HK2PR06MB0802F1757D01551310AB832EB7790%40HK2PR06MB0802.apcprd06.prod.outlook.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
杨振军(Zhenjun Yang)
Dear Cath,
Thank you for your answer.
After I click on your link, I can see the animo acid sequence, however, when I enter a new gene or new region, it still show the DNA sequence, and I always set the section“Codon Translation”to "Use default species reading frames for translation".
Minutes later, I tried to set the section“Codon Translation”to "No codon translation" and submit, and then, I set it back to "Use default species reading frames for translation" and submit,
Finally, I saw the animo acid sequence of my gene!
I don’t know if it is a bug of UCSC, but I still deeply appreciate your answer.
Thank you and best regards.
Yours sincerely,
Zhenjun Yang
发件人: Cath Tyner [mailto:ca...@ucsc.edu]
发送时间: 2017年9月30日
8:32
收件人: 杨振军(Zhenjun Yang) <yangz...@genomics.cn>
抄送: gen...@soe.ucsc.edu
主题: Re: [genome] Vertebrate Multiz Alignment & Conservation (46 Species)
Hello,
Thank you for contacting the UCSC Genome Browser support team. From the track setting page, you can select an option to show codon translation. For example:
1. Click on the link to see this "hg19 session":http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=cath&hgS_otherUserSessionName=MLQ20241.
2. Scroll down to the "Cons 46-Way" track and click on the name underlined in blue to go to the track settings page. Alternatively, you can also right-click on the left vertical gray button to the left of the track in the browser graphic.
3. From the track's settings page: http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=cons46way, look in the yellow box with the tree, "Multiz Alignments Configuration."
4. Under the section "Codon Translation," make sure you have selected, "Use default species reading frames for translation" instead of "No codon translation."
Please respond to this list if you have further questions!
Thank you for contacting the UCSC Genome Browser support team.
Please send new and follow-up questions to one of our UCSC Genome Browser mailing lists below:
* Confidential/private help: Email
UCSC Genome Browser Announcements List (email alerts for new data & software):
* Subscribe: Email genome-annou...@soe.ucsc.edu
* Unsubscribe: Email genome-announ...@soe.ucsc.edu
Jairo Navarro Gonzalez
Hello Zhenjun,
Thank you for using the UCSC Genome Browser and your follow-up question.
I was unable to replicate your issue. If your browser isn't allowing cookies, this will result in your settings not being saved.
If you are allowing cookies and the problem arises again, you can change your URL from "http://genome.ucsc.edu/cgi-bin/hgTracks...." to "http://genome.ucsc.edu/cgi-bin/cartDump" and you your screen will show settings that might give us clues to what is going on. You can send a copy of this information as a text file off-list directly to me.
I hope this is helpful. If you have any further questions, please reply to gen...@soe.ucsc.edu.
All messages sent to that address are archived on a publicly-accessible Google Groups forum.
If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
Jairo Navarro
UCSC Genomics Institute
* Subscribe: Email genome-announce+subs...@soe.ucsc.edu
* Unsubscribe: Email genome-announce+unsub...@soe.ucsc.edu
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/HK2PR06MB08022A8B57DE67C11D2C7E30B77F0%40HK2PR06MB0802.apcprd06.prod.outlook.com.
