Gerritsen, M (onco)
The contents of this message are confidential and only intended for the eyes of the addressee(s). Others than the addressee(s) are not allowed to use this message, to make it public or to distribute or multiply this message in any way. The UMCG cannot be held responsible for incomplete reception or delay of this transferred message.
Brian Lee
Dear Mylene,
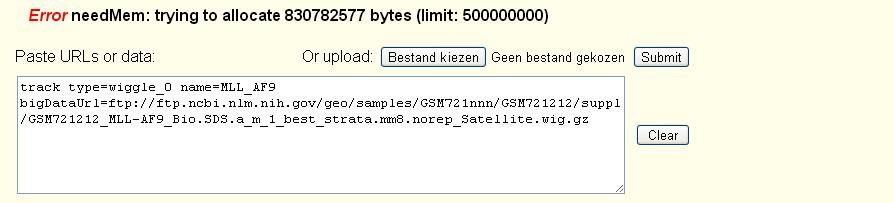
Thank you for using the UCSC Genome Browser and your question about the memory limit error you are seeing.
You are correct that the path to take is to make a bigWig out of such files. To do so you would need the properly compiled wigToBigWig utility for your system. You can do "uname -a" and pick the utility from the matching directory here: http://hgdownload.soe.ucsc.edu/admin/exe/
Once you download the wigToBigWig utility you would also need to get the chrom.sizes file for the matching assembly, in this case mm8. You can obtain it here: http://hgdownload.cse.ucsc.edu/goldenPath/mm8/bigZips/mm8.chrom.sizes
Then one would normally run the utility with this command on the obtained unzipped wig file:wigToBigWig GSM721212.wig mm8.chrom.sizes GSM721212.bigWig
However, this file has some errors, the first of which is there are some annotations that extend past the end of chromosomes, so you can avoid that problem using the -clip option, to clip these final entries:wigToBigWig -clip GSM721212.wig mm8.chrom.sizes GSM721212.bigWig
The file also has some errors where "start=0" needs to be changed to "start=1", an example is line 86094512 for chrM: fixedStep chrom=chrM start=0 step=25 span=25
Please take these steps yourself for future files, but for your convenience I put these together here: http://hgwdev.cse.ucsc.edu/~brianlee/customTracks/GSM721212.txt
These tracks can be loaded with this URL: http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&hgt.customText=http://hgwdev.cse.ucsc.edu/~brianlee/customTracks/GSM721212.txt
Note that you can upload sections of the wig file, if for example you were only interested in chrM data, you could upload just that section and avoid the memory problem. However, the best solution is to create a bigWig file and host it in a publicly accessible directory. More about bigWig files can be found here: http://genome.ucsc.edu/FAQ/FAQformat.html#format6.1
Thank you again for your inquiry and using the UCSC Genome Browser. If you have any further questions, please reply to gen...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
All the best,
Brian Lee
UCSC Genome Bioinformatics Group
--
