Extracting data from the table browser
Samarakkody, Ann
Hello,
I'm trying to extract data from the table browser and I was hoping to get help from you in that regards.
I want to extract data from the human data base for the following.
1. genomic intervals for all exons
2. Genomic intervals for all introns
3. Specific region (-500 to +500 from TSS) of promoter for all genes
I tried to address number 1 by using the exon start and end option on the table browser (screen shots attached below) however I'm unable to extract the interval from the output file. What would be the best way to extract the data for 1-3?
Any suggestions or directions would be great appreciated!
Thank you
Sincerely,
Ann Sanoji Samarakkody
PhD candidate - Nechaev Lab
Program of Anatomy and Cell Biology
Department of Biomedical Sciences
School of Medicine and Health Sciences
University of North Dakota
Jairo Navarro Gonzalez
To obtain Genomic Intervals for Exons and Introns
Step 1: Configure Table Browser Settings
Clade: Mammal
Genome: Human
Assembly: Feb. 2009 (GRCh37/hg19)
Group: Genes and Gene Predictions
Track: UCSC Genes
Region: genome
Output format: BED - browser extensible data
Step 2: Click 'get output'
Select either 'Exons plus' or 'Introns plus'
Select 'get BED'
To obtain specific region of promoter for all genes
Step 1: Configure Table Browser Settings
Clade: Mammal
Genome: Human
Assembly: Feb. 2009 (GRCh37/hg19)
Group: Genes and Gene Predictions
Track: UCSC Genes
Region: genome
Output format: selected fields from primary and related tables
Output file: enter a name for your file
select 'get ouput'
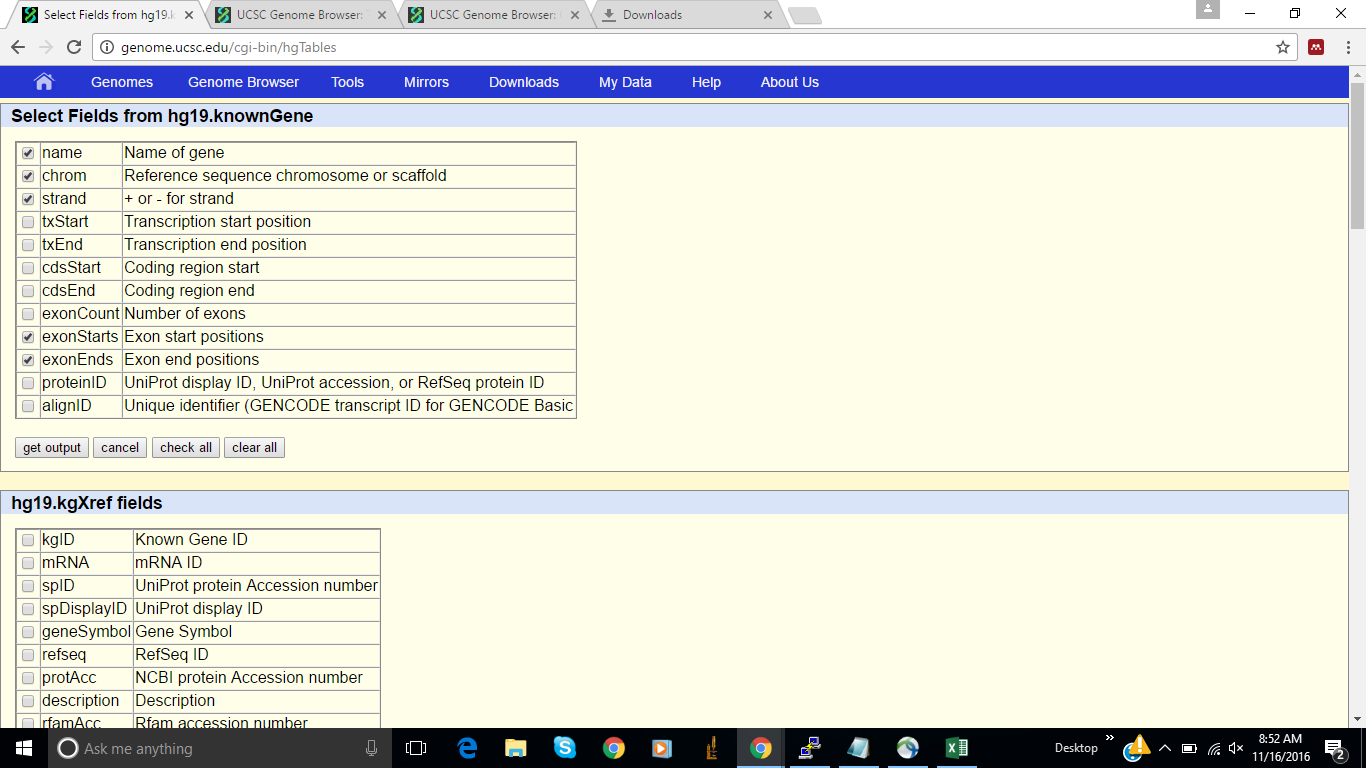
Step 2: Choose output fields
From 'Select Fields from hg19.knownGene', select name, chrom, and txStart
click 'get output'
Step 3: Use a script to change the output file
You should have an output like:
Using a script, copy and add another column to each line of the txStart siteThe result should look something like:
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser discussion list" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
Jairo Navarro Gonzalez
Clade: MammalGenome: HumanAssembly: Feb. 2009 (GRCh37/hg19)Group: Genes and Gene PredictionsTrack: UCSC GenesRegion: genome
filter: changes depending on the strand you are retrieving
- For the positive (+) strand, your filter will be: strand does match +
- For the negative (-) strand, your filter will be: strand does match -
Output format: selected fields from primary and related tablesOutput file: enter a name for your file
select 'get output'
From 'Select Fields from hg19.knownGene', select name, chrom
If you are filtering for the positive strand, choose txStartIf you are filtering for the negative strand, choose txEndclick 'get output'
