Table browser
5 views
Skip to first unread message
Jennifer Hauenstein
Dec 20, 2017, 11:21:25 AM12/20/17
to gen...@soe.ucsc.edu
Good morning~
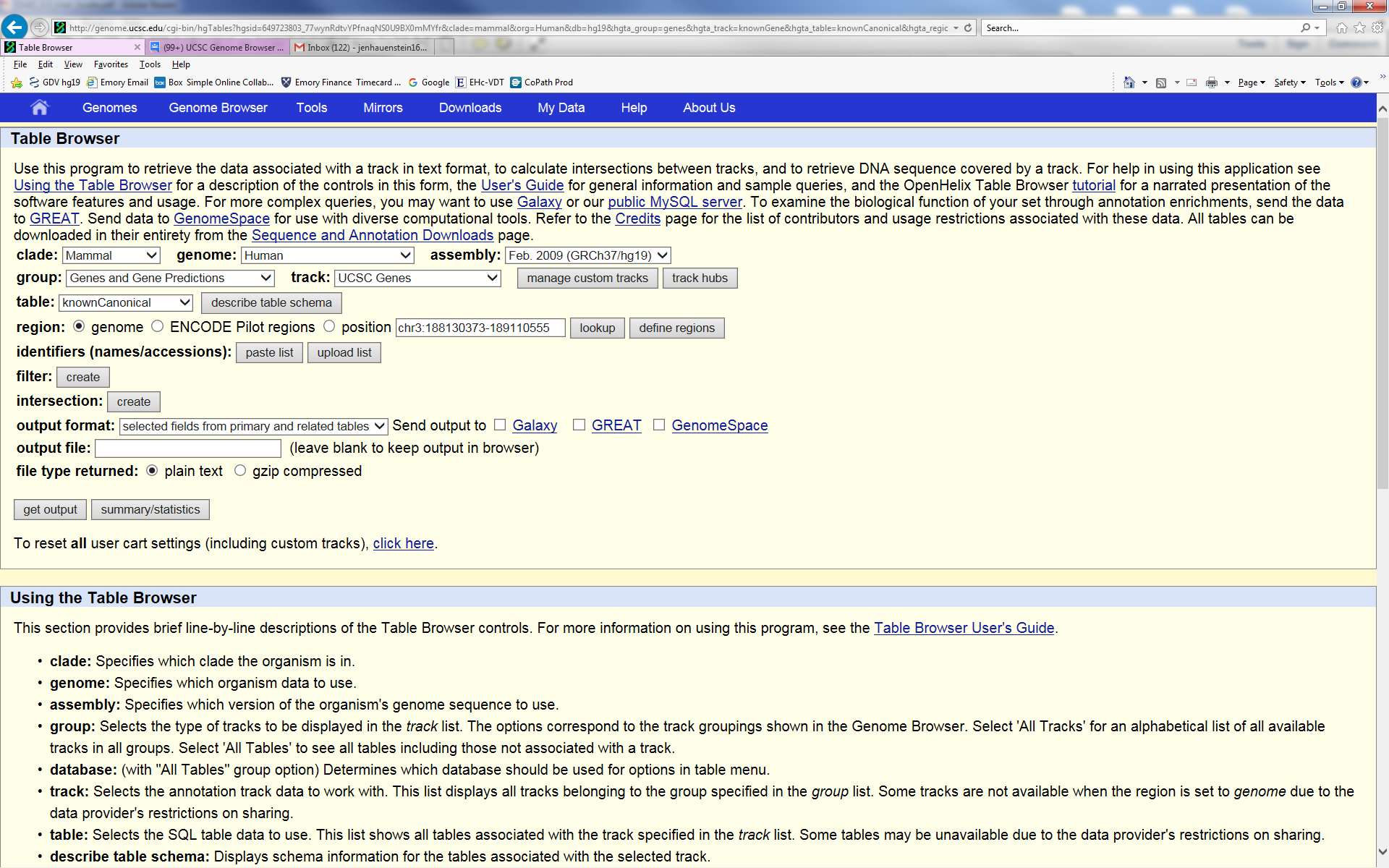
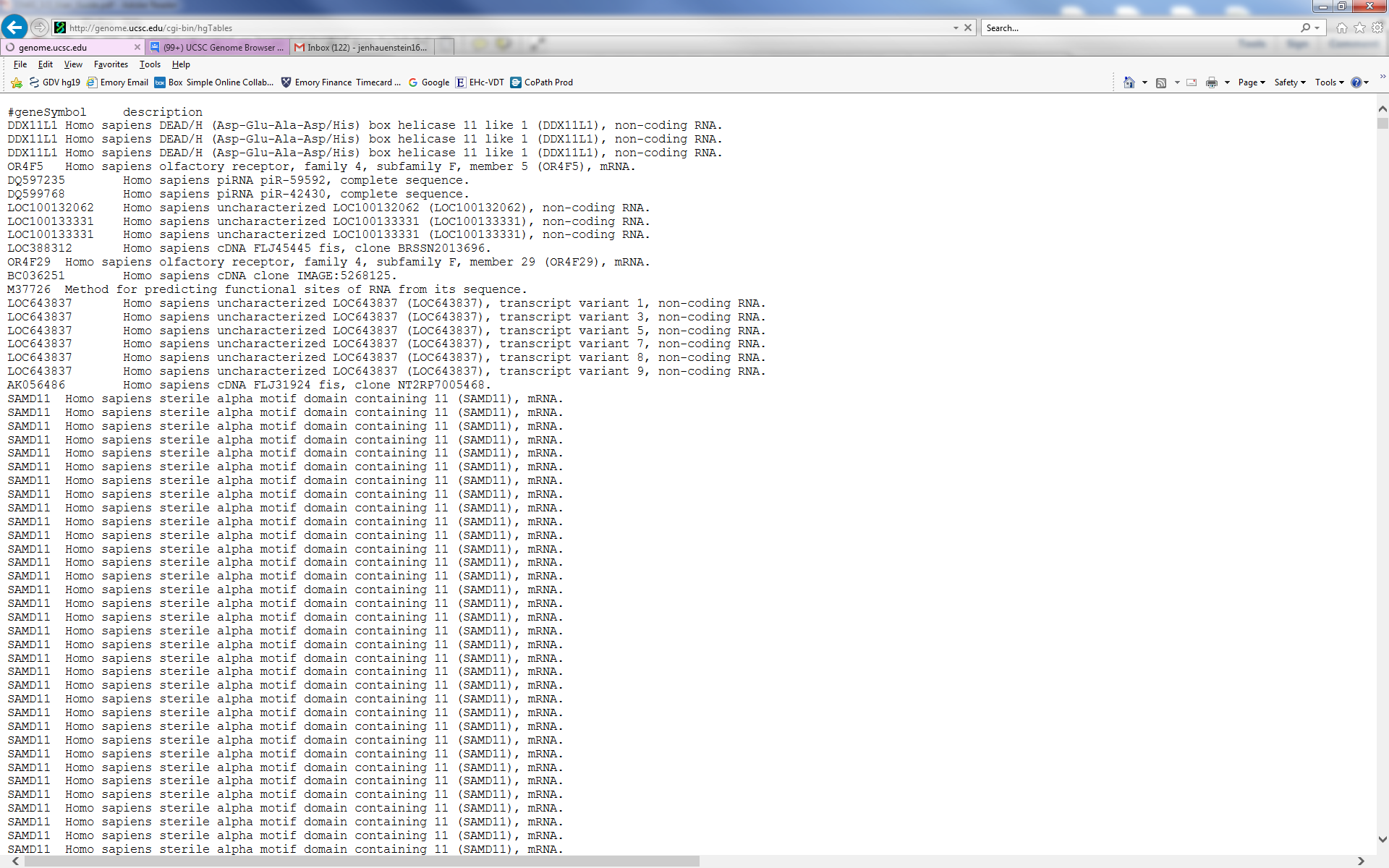
I would like to use the table browser to create a list of genes, their OMIM numbers, and their short descriptions from a segment that I viewed in the genome browser. From the selections I chose, I keep getting multiple entries for each gene. Here are the selections I am normally choosing that give me the multiple gene output. Also, I would like to get the OMIM numbers but that track is greyed out. Which selections can I choose to get a single output of the gene name, description, and OMIM number for each gene in the segment?
Thanks~
Jen
Robert Kuhn
Dec 20, 2017, 4:10:37 PM12/20/17
to Jennifer Hauenstein, UCSC Genome Browser Mailing List, Robert Kuhn
Hello, Jennifer,
Thanks for your question. The OMIM tables are not accessible via the Table
Browser if you have "genome" selected as a region, per our agreement with
the OMIM team not to redistribute their entire dataset. The DDX11L1 gene
in your output is the first gene on chr1 and your output is likely very large
(whole genome)? Choose the button next to "position" to restrict your query
to the chr3 region you were viewing on the Browser. The OMIM tables should
then be available to you.
The multiple entries are due to the fact that the knownGene table will give a result
for each isoform. If you wish one isoform per locus, use the knownCannonical
table in the UCSC Genes track. Though I see in your image that you already
have that table selected, your output shows that the query you executed for that
particular output had the knownGene table selected.
The following video shows most of the process you request:
You may also be interested in the Data Integrator, which can do what the
Table Browser does but also a bit more. Here is a short video demonstrating
almost the exact problem you describe, including obtaining OMIM data:
Best wishes, and thanks for using the Genome Browser.
--b0b kuhn
ucsc genome bioinformatics group
Browser training workshops available via: http://genome.ucsc.edu/training
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To post to this group, send email to gen...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/CAE%2BTxMgTQhrXcRjYLY%2Bqj2V9TTrygR2ESP9reUMktfSPLgPBig%40mail.gmail.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
Reply all
Reply to author
Forward
0 new messages
