HELP: GBiC local mirror only shows my first downloaded genome
Zhibo Ma
Hi,
I installed a UCSC browser using the GBiC browserSetup.sh script. And I have downloaded the dm6 genome assembly. Everything worked fine until I downloaded a few more genomes using the mirror tool. The downloading/mirror process was successful. But after it’s completed, the localhost browser still only shows the dm6 genome. None of the other drosophila species genome that I have downloaded is shown. I noticed that the represented species section is missing, no matter it’s in the local only or on-the-fly mode. (see the two screenshots, first is local and the second is on-the-fly).
Same thing happened to a few genome databases of other drosophila species that I added. I do not see them in the listed under fruitfly. I thought it might because I didn’t add it correctly. Now I feel it might due to the same error somewhere.
Could someone help me figure out which part is wrong and how could I show all the genomes that I downloaded with the browserSetup.sh mirror ?
Thank you!
Zhibo
Below is a screenshot when I turned the on-the-fly mode off, which only shows the dm6 genome. No represented species box:
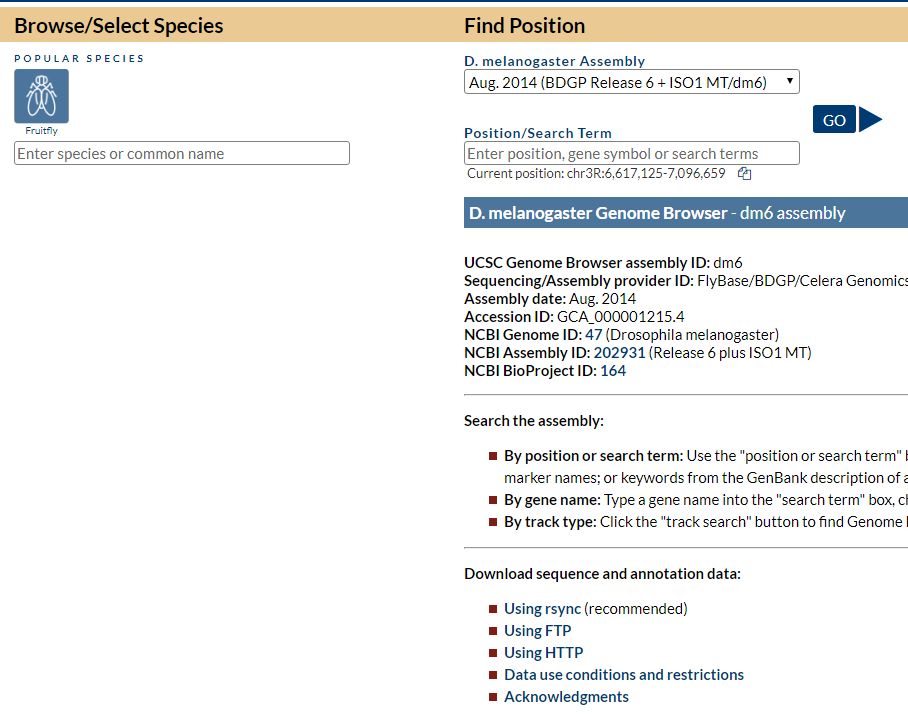
This one below is when I turned the on-the-fly mode on, which shows all the popular species. But there is no “represented species” box to select a species:
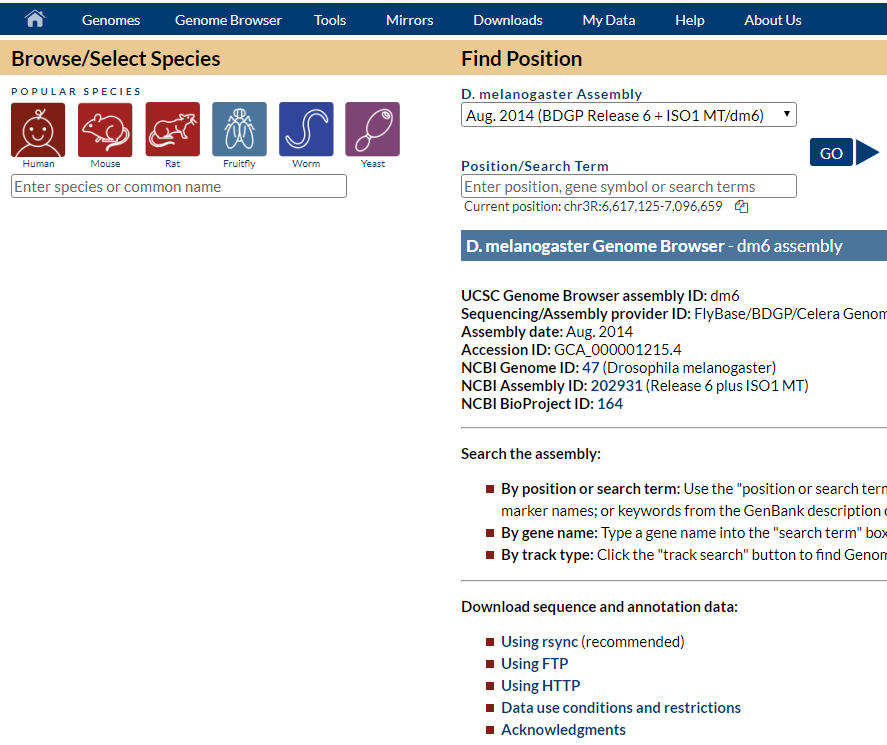
Zhibo Ma
Graduate student
Department of Cellular Biology
University of Georgia
Athens, GA 30605
Brian Lee
- Do all of the locally installed databases have active=1 in the local hgcentral.dbDb table?
- Does the file /usr/local/apache/htdocs/js/dbDbTaxonomy.js exist?
- If so, what is it set to? Is the file readable by apache?
- If not does hg.conf include the setting hgGateway.dbDbTaxonomy?
dbDbTree is found and has at least one active species. When everything is working right:- hgGateway.c finds a JS file that defines
dbDbTreeand includes it as a<script>in the page HTML. - hgGateway.c also defines a JS object
activeGenomesby querying hgcentral.dbDb to get dbs with active=1. - hgGateway.js gets
dbDbTreeandactiveGenomes, and prunes everything indbDbTreethat is not inactiveGenomes. - If @dbDbTree is non-empty after all that, then hgGateway.js draws the tree.
By default, the JS file that defines dbDbTree is ../js/dbDbTaxonomy.js (i.e. /usr/local/apache/htdocs/js/dbDbTaxonomy.js). A different path, relative to cgi-bin, can be defined in hg.conf like this:
On our site and in our GBiC/GBiB's the /usr/local/apache/htdocs/js/dbDbTaxonomy.js file exists and hgcentral.dbDb has lots of databases with active=1. If that's the case for you too, then it would be helpful if you would check the console output from Developer Tools to share with us.Thank you again for your inquiry and using the UCSC Genome Browser. If you have any further public questions, please reply to genome...@soe.ucsc.edu. All messages sent to that address are archived on a publicly-accessible forum. If your question includes sensitive data, you may send it instead to genom...@soe.ucsc.edu.
All the best,
Brian Lee
UC Santa Cruz Genomics Institute
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Mirror-Specific Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome-mirror+unsubscribe@soe.ucsc.edu.
To post to this group, send email to genome...@soe.ucsc.edu.
Visit this group at https://groups.google.com/a/soe.ucsc.edu/group/genome-mirror/.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome-mirror/SN1PR0201MB1792492984472FF0A229E067C8840%40SN1PR0201MB1792.namprd02.prod.outlook.com.
For more options, visit https://groups.google.com/a/soe.ucsc.edu/d/optout.
Zhibo Ma
Dear Brian,
To your requested info:
- Do all of the locally installed databases have active=1 in the local hgcentral.dbDb table?
Yes. All the 5 databases(4 from mirror download, 1 from custom addition) have active=1 in the local hgcentral.dbDb
I also noticed that total of 195/295 genome databases in dbDb are active=1, although I did not download those genomes.
mysql> select * from dbDb where name="dm6" OR name="droPer1" OR name="droSim1" OR name="droSec1" OR name="droVir3";
+---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
| name | description | nibPath | organism | defaultPos | active | orderKey | genome | scientificName | htmlPath | hgNearOk | hgPbOk | sourceName | taxId |
+---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
| dm6 | Aug. 2014 (BDGP Release 6 + ISO1 MT/dm6) | /gbdb/dm6 | D. melanogaster | chr2L:826,001-851,000 | 1 | 4067 | D. melanogaster | Drosophila melanogaster | /gbdb/dm6/html/description.html | 0 | 0 | The FlyBase Consortium/Berkeley Drosophila Genome Project/Celera Genomics | 7227 |
| droSim1 | Apr. 2005 (WUGSC mosaic 1.0/droSim1) | /gbdb/droSim1 | D. simulans | chr2L:827,150-853,526 | 1 | 4110 | D. simulans | Drosophila simulans | /gbdb/droSim1/html/description.html | 0 | 0 | WUSTL version 1.0 | 7240 |
| droPer1 | Oct. 2005 (Broad/droPer1) | /gbdb/droPer1 | D. persimilis | super_1:7109001-7125000 | 1 | 4085 | D. persimilis | Drosophila persimilis | /gbdb/droPer1/html/description.html | 0 | 0 | Broad 28 Oct. 2005 | 7234 |
| droSec1 | Oct. 2005 (Broad/droSec1) | /gbdb/droSec1 | D. sechellia | super_14:795001-815000 | 1 | 4105 | D. sechellia | Drosophila sechellia | /gbdb/droSec1/html/description.html | 0 | 0 | Broad 28 Oct. 2005 | 7238 |
| droVir3 | Feb. 2006 (Agencourt CAF1/droVir3) | /gbdb/droVir3 | D. virilis | scaffold_1:1-1000 | 1 | 4123 | D. virilis | Drosophila virilis | /gbdb/droVir3/html/description.html | 0 | 0 | Agencourt CAF1 | 7244 |
+---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
5 rows in set (0.00 sec)
- Does the file /usr/local/apache/htdocs/js/dbDbTaxonomy.js exist?
Yes.
- If so, what is it set to? Is the file readable by apache?
- Not sure what are you asking. Below has the info I think you might asked for:
$ls -al dbDbTaxonomy.js
-rw-rw-r-- 1 mzhibo mzhibo 96457 May 7 17:43 dbDbTaxonomy.js
$head dbDbTaxonomy.js
/* jshint maxlen: false, unused: false */
var dbDbTree = ["root", 1, null,
[ ["cellular organisms", 131567, null,
[ ["Eukaryota", 2759, null,
[ ["Opisthokonta", 33154, null,
[ ["Metazoa", 33208, null,
[ ["Eumetazoa", 6072, null,
[ ["Bilateria", 33213, null,
[ ["Deuterostomia", 33511, null,
[ ["Chordata", 7711, null,
My hg.conf file does not include setting of
hgGateway.dbDbTaxonomy. But it made no difference after I added
hgGateway.dbDbTaxonomy=/usr/local/apache/htdocs/js/dbDbTaxonomy.js
By the way, although there is no represented tree, I can search and use the genomes that I have mirror downloaded or added.
The represented tree will comeback when the mirror is set to on-the-fly mode.
Thanks for the help.
Zhibo
--
To unsubscribe from this group and stop receiving emails from it, send an email to genome-mirro...@soe.ucsc.edu.
Brian Lee
Can you check what the console is displaying on your browser's developer tools when you are on the hgGateway page, it might provide us clues to what is happening.
Also, just to verify, are you able to access the track display for these assemblies, for example navigate to the individual Genome Browser page and see your data /cgi-bin/hgTracks?db=xxxx such as /cgi-bin/hgTracks?db=droSec1)?
Brian
--
To unsubscribe from this group and stop receiving emails from it, send an email to genome-mirror+unsubscribe@soe.ucsc.edu.
Zhibo Ma
Hi Brian,
It looks like the hgGateway did not find the js file as indicated in the hg.conf file.

In my hg.conf file
hgGateway.dbDbTaxonomy=/usr/local/apache/htdocs/js/dbDbTaxonomy.js
I also tried
hgGateway.dbDbTaxonomy=../js/dbDbTaxonomy.js
None of help.
Thanks!
Zhibo
From: Brian Lee <bria...@soe.ucsc.edu>
Sent: Tuesday, May 8, 2018 7:06 PM
To: Zhibo Ma <mzh...@uga.edu>
Subject: Re: [genome-mirror] HELP: GBiC local mirror only shows my first downloaded genome
Hi Zhibo,
Thanks for sharing that the track pages are working fine. Could you open the developer tools for the internet browser you might be using (or provide a URL to your mirror if it is accessible to the outside). For example, if you were using Chrome as a browser, you would go to the view menu and then select the Developer menu and then Developer Tools. If you then click the "Console" option you should see error reports that might help us debug what is occurring. If you are using Firefox the Developer menu is under Tools and then you can select console.
All the best,
Brian
On Tue, May 8, 2018 at 3:49 PM, Zhibo Ma <mzh...@uga.edu> wrote:
Dear Brian,
The individual Genome Browser page displays all the loaded tracks of that genome correctly. The problem seems to be only missing the represented tree.
About the broswer’s developer tools. Where can I find it?
Thanks,
--
To unsubscribe from this group and stop receiving emails from it, send an email to genome-mirro...@soe.ucsc.edu.
Hiram Clawson
On 5/8/18 4:42 PM, Zhibo Ma wrote:
> Hi Brian,
> It looks like the hgGateway did not find the js file as indicated in the hg.conf file.
> In my hg.conf file
> hgGateway.dbDbTaxonomy=/usr/local/apache/htdocs/js/dbDbTaxonomy.js
> I also tried
> hgGateway.dbDbTaxonomy=../js/dbDbTaxonomy.js
> None of help.
>
> Thanks!
> Zhibo
>
> From: Brian Lee <bria...@soe.ucsc.edu>
> Sent: Tuesday, May 8, 2018 7:06 PM
> To: Zhibo Ma <mzh...@uga.edu>
> Subject: Re: [genome-mirror] HELP: GBiC local mirror only shows my first downloaded genome
>
> Hi Zhibo,
>
> Thanks for sharing that the track pages are working fine. Could you open the developer tools for the internet browser you might be using (or provide a URL to your mirror if it is accessible to the outside). For example, if you were using Chrome as a browser, you would go to the view menu and then select the Developer menu and then Developer Tools. If you then click the "Console" option you should see error reports that might help us debug what is occurring. If you are using Firefox the Developer menu is under Tools and then you can select console.
>
> All the best,
> Brian
>
> Dear Brian,
> The individual Genome Browser page displays all the loaded tracks of that genome correctly. The problem seems to be only missing the represented tree.
> About the broswer’s developer tools. Where can I find it?
> Thanks,
> Zhibo
>
>
> From: Brian Lee <bria...@soe.ucsc.edu<mailto:bria...@soe.ucsc.edu>>
> Sent: Tuesday, May 8, 2018 6:35 PM
>
> Cc: genome...@soe.ucsc.edu<mailto:genome...@soe.ucsc.edu>
>
> Dear Zhibo,
>
> Can you check what the console is displaying on your browser's developer tools when you are on the hgGateway page, it might provide us clues to what is happening.
>
> Also, just to verify, are you able to access the track display for these assemblies, for example navigate to the individual Genome Browser page and see your data /cgi-bin/hgTracks?db=xxxx such as /cgi-bin/hgTracks?db=droSec1)?
>
> Brian
>
> Dear Brian,
> To your requested info:
>
> I also noticed that total of 195/295 genome databases in dbDb are active=1, although I did not download those genomes.
> mysql> select * from dbDb where name="dm6" OR name="droPer1" OR name="droSim1" OR name="droSec1" OR name="droVir3";
> +---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
> | name | description | nibPath | organism | defaultPos | active | orderKey | genome | scientificName | htmlPath | hgNearOk | hgPbOk | sourceName | taxId |
> +---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
> | dm6 | Aug. 2014 (BDGP Release 6 + ISO1 MT/dm6) | /gbdb/dm6 | D. melanogaster | chr2L:826,001-851,000 | 1 | 4067 | D. melanogaster | Drosophila melanogaster | /gbdb/dm6/html/description.html | 0 | 0 | The FlyBase Consortium/Berkeley Drosophila Genome Project/Celera Genomics | 7227 |
> | droSim1 | Apr. 2005 (WUGSC mosaic 1.0/droSim1) | /gbdb/droSim1 | D. simulans | chr2L:827,150-853,526 | 1 | 4110 | D. simulans | Drosophila simulans | /gbdb/droSim1/html/description.html | 0 | 0 | WUSTL version 1.0 | 7240 |
> | droPer1 | Oct. 2005 (Broad/droPer1) | /gbdb/droPer1 | D. persimilis | super_1:7109001-7125000 | 1 | 4085 | D. persimilis | Drosophila persimilis | /gbdb/droPer1/html/description.html | 0 | 0 | Broad 28 Oct. 2005 | 7234 |
> | droSec1 | Oct. 2005 (Broad/droSec1) | /gbdb/droSec1 | D. sechellia | super_14:795001-815000 | 1 | 4105 | D. sechellia | Drosophila sechellia | /gbdb/droSec1/html/description.html | 0 | 0 | Broad 28 Oct. 2005 | 7238 |
> | droVir3 | Feb. 2006 (Agencourt CAF1/droVir3) | /gbdb/droVir3 | D. virilis | scaffold_1:1-1000 | 1 | 4123 | D. virilis | Drosophila virilis | /gbdb/droVir3/html/description.html | 0 | 0 | Agencourt CAF1 | 7244 |
> +---------+------------------------------------------+---------------+-----------------+-------------------------+--------+----------+-----------------+-------------------------+-------------------------------------+----------+--------+---------------------------------------------------------------------------+-------+
> 5 rows in set (0.00 sec)
>
> Yes.
>
> * If so, what is it set to? Is the file readable by apache?
>
> * Not sure what are you asking. Below has the info I think you might asked for:
> * Does the file /usr/local/apache/htdocs/js/dbDbTaxonomy.js exist?
>
> * If so, what is it set to? Is the file readable by apache?
> * If not does hg.conf include the setting hgGateway.dbDbTaxonomy?
>
> * hgGateway.c also defines a JS object activeGenomes by querying hgcentral.dbDb to get dbs with active=1.
> * hgGateway.js gets dbDbTree and activeGenomes, and prunes everything in dbDbTree that is not in activeGenomes.
> * If @dbDbTree is non-empty after all that, then hgGateway.js draws the tree.
> By default, the JS file that defines dbDbTree is ../js/dbDbTaxonomy.js (i.e. /usr/local/apache/htdocs/js/dbDbTaxonomy.js). A different path, relative to cgi-bin, can be defined in hg.conf like this:
>
> hgGateway.dbDbTaxonomy=../js/myLocalTaxonomy.js
> On our site and in our GBiC/GBiB's the /usr/local/apache/htdocs/js/dbDbTaxonomy.js file exists and hgcentral.dbDb has lots of databases with active=1. If that's the case for you too, then it would be helpful if you would check the console output from Developer Tools to share with us.
>
> All the best,
>
> Brian Lee
>
> UC Santa Cruz Genomics Institute
>
>
>
>
>
>
> On Sat, May 5, 2018 at 7:19 PM, Zhibo Ma <mzh...@uga.edu<mailto:mzh...@uga.edu>> wrote:
> Hi,
> I installed a UCSC browser using the GBiC browserSetup.sh script. And I have downloaded the dm6 genome assembly. Everything worked fine until I downloaded a few more genomes using the mirror tool. The downloading/mirror process was successful. But after it’s completed, the localhost browser still only shows the dm6 genome. None of the other drosophila species genome that I have downloaded is shown. I noticed that the represented species section is missing, no matter it’s in the local only or on-the-fly mode. (see the two screenshots, first is local and the second is on-the-fly).
> Same thing happened to a few genome databases of other drosophila species that I added. I do not see them in the listed under fruitfly. I thought it might because I didn’t add it correctly. Now I feel it might due to the same error somewhere.
> Could someone help me figure out which part is wrong and how could I show all the genomes that I downloaded with the browserSetup.sh mirror ?
> Thank you!
> Zhibo
> Below is a screenshot when I turned the on-the-fly mode off, which only shows the dm6 genome. No represented species box:
>
> Graduate student
> Department of Cellular Biology
> University of Georgia
> Athens, GA 30605
>
> --
>
> ---
> You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Mirror-Specific Support" group.
> To post to this group, send email to genome...@soe.ucsc.edu<mailto:genome...@soe.ucsc.edu>.
Zhibo Ma
Problem is solved.
I found that there is another hg.conf file in the cgi-bin directory. I was adding the "hgGateway.dbDbTaxonomy=../js/dbDbTaxonomy.js" line to the ~/.hg.conf file earlier.
After I added this line to the /usr/local/apache/cgi-bin/hg.conf, the problem is solved!
Thanks for your help!
Zhibo
