Read count track for Tabula Muris dataset
Manjegowda, Mohan C (mcm3ha)
Mohan C Manjegowda, PhD
Dept. of Pharmacology
University of Virginia
Charlottesville, VA-22908
Luis Nassar
Hello, Mohan.
Thank you for your interest in the Browser.
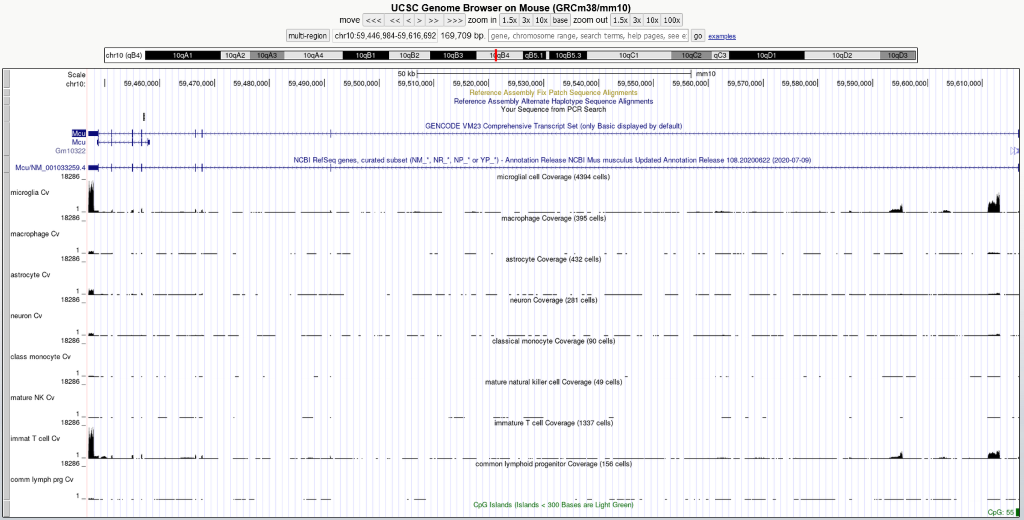
While we can confirm that what you are pointing out here is peculiar and that the data as we display it is correct, we recommend you contact Tabula Muris directly to hear if they have a better explanation of what is occurring.
As you share, we see peaks outside the exon boundaries (https://genome.ucsc.edu/s/Lou/RM30509.1). One of our engineers points out that according to the Tabula Muris paper they used STAR aligner, which uses exon information but does not align to just exons. You could try contacting Angela Pisco (angela...@czbiohub.org) who is listed as one of the contacts for the project (https://data.humancellatlas.org/explore/projects/e0009214-c0a0-4a7b-96e2-d6a83e966ce0). If you do decide to contact them, consider cc'ing my colleague Max (mhae...@ucsc.edu) as he brought this data into the Browser and is interested to see what they have to say.
I hope this is helpful. Please include gen...@soe.ucsc.edu in any replies to ensure visibility by the team. All messages sent to that address are archived on our public forum. If your question includes sensitive information, you may send it instead to genom...@soe.ucsc.edu.
Lou Nassar
UCSC Genomics Institute
--
---
You received this message because you are subscribed to the Google Groups "UCSC Genome Browser Public Support" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genome+un...@soe.ucsc.edu.
To view this discussion on the web visit https://groups.google.com/a/soe.ucsc.edu/d/msgid/genome/MN2PR13MB358140732597EAD27F22DAF8FCC69%40MN2PR13MB3581.namprd13.prod.outlook.com.
