Unable to add to User-created Annotations
19 views
Skip to first unread message
M
Mar 18, 2021, 5:11:33 AM3/18/21
to apollo
My lab is currently running Apollo v2.3.1 on Debian 10 with Tomcat8. We recently had to reboot the system, and since then we have been unable to add any features to the User-created Annotations track, either by drag-and-drop or by right-click. The feature works for neither the new organism I added since the reboot, nor the existing ones for which it worked previously.
I'm unsure of whether this is relevant or offers any clues, but when I attempt to add a feature to User-created Annotations, the file localhost_access_log.<date>.txt updates with:
"POST /apollo/sequence/setCurrentSequenceLocation/?name=Sm_000000F&start=186935&end=200351&suppressOutput=true&clientToken=2102888520838143417648978935 HTTP/1.1" 200 12
"POST /apollo/sequence/setCurrentSequenceLocation/?name=Sm_000000F&start=186935&end=200351&suppressOutput=true&clientToken=2102888520838143417648978935 HTTP/1.1" 200 12
When I log into the browser, the catalina.out file also notes:
WARN apollo.PreferenceService - No user present, so using the client token
ERROR apollo.PermissionService - Username not supplied so can not authenticate.
WARN apollo.PermissionService - Failed to authenticate user
ERROR apollo.PermissionService - Username not supplied so can not authenticate.
WARN apollo.PermissionService - Failed to authenticate user
I'm still able to log in and view the genome as normal, however, so I'm unsure if my problem is related to some kind of permissions issue.
Any guidance you can give would be greatly appreciated.
Kind regards,
Matt
Nathan Dunn
Mar 18, 2021, 10:27:19 AM3/18/21
to M, apollo
My guess is that you have a Canvas Track configured. https://genomearchitect.readthedocs.io/en/latest/Data_loading.html?highlight=canvas#canvas-vs-html-in-apollo
Can you upload a screen shot?
Also, if you have track list information that would be helpful.
Cheers,
Nathan
Nathan Dunn
Mar 18, 2021, 11:23:35 AM3/18/21
to M, apollo
This is the right information and it looks ike it is setup correctly.
If you are using the admin user (you should see the organism panel, etc.) what do you see in the javascript console and network tabs when you attempt to create an annotation as well as when you first load?
If you are unfamiliar with using the javascript console in Chrome (though any browser is fine):
Cheers,
Nathan
On Mar 18, 2021, at 7:49 AM, M <matt_p...@hotmail.com> wrote:
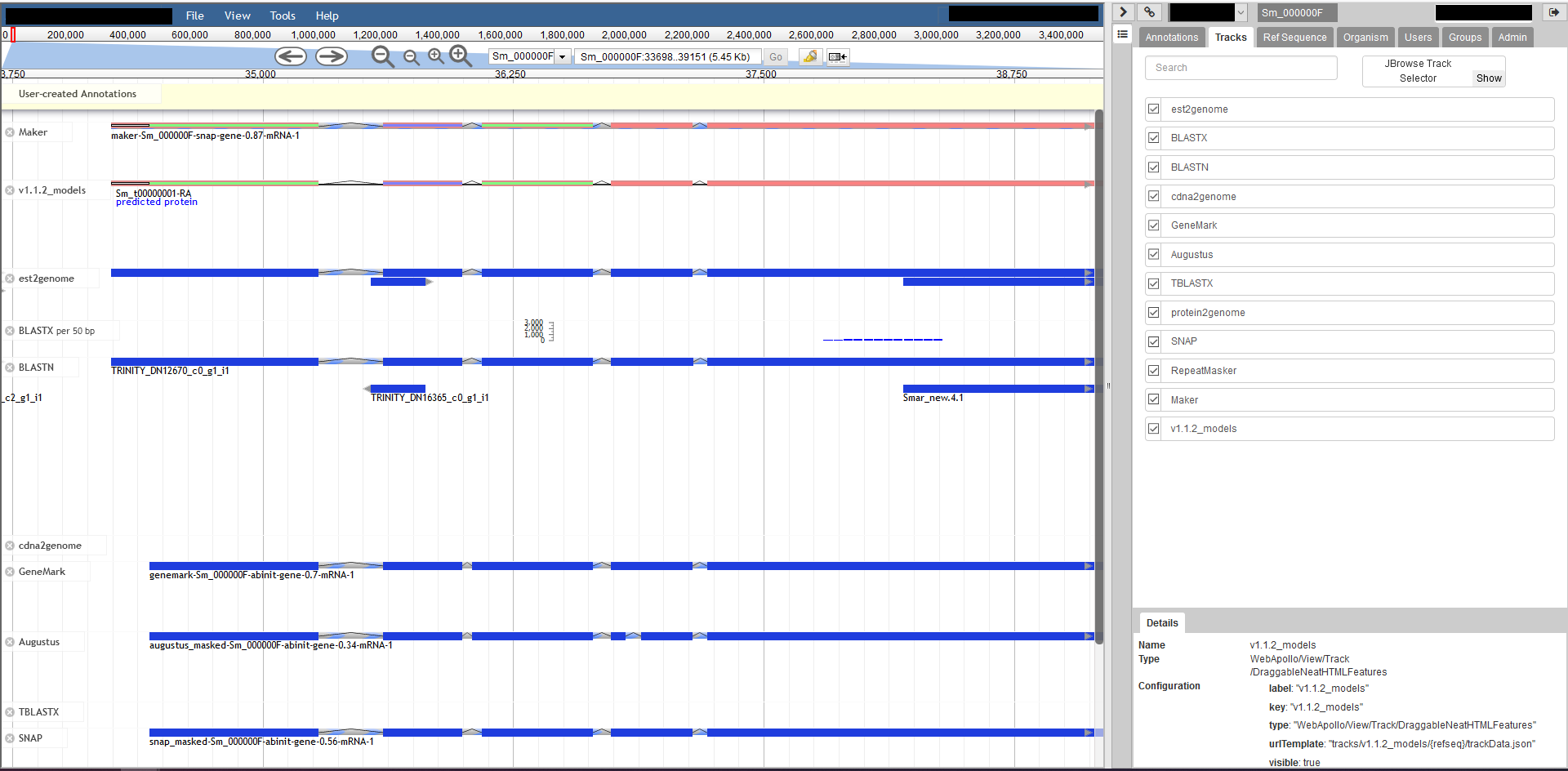
I've attached a screenshot below, as well as the trackList.json file. I hope I understood correctly which information you were after.Many thanks,Matt
--
To unsubscribe from this group and stop receiving emails from it, send an email to apollo+un...@lbl.gov.
<trackList.json><WebApolloScreenshot.png>
M
Mar 18, 2021, 11:43:47 AM3/18/21
to apollo, Nathan Dunn, apollo, M
Here's the console output from loading the page and trying to add a feature. Drag-and-drop of features doesn't cause a response in the console, whereas 'right-click > Create new annotation > gene' gives the 'biotype not found' message on the bottom line.
Many thanks,
Matt
Nathan Dunn
Mar 18, 2021, 4:36:27 PM3/18/21
to M, apollo
That message is more informative than an error in this case.
Probably the quickest way for me to help resolve it is for you to send (from easiest to hardest):
- some sample genes from your GFF3
- create a small zip file for one chromosome and one gff or just your entire jbrowse directory if it is small (< 30 MBs).
Feel free to email me directly or send to the list.
Nathan
<console.18-03-2021.log>
Reply all
Reply to author
Forward
0 new messages