Get gene sequence
3 views
Skip to first unread message
Michał T. Lorenc
Mar 8, 2022, 11:58:37 PM3/8/22
to apollo
Hi all,
Apollo allows to drag-n-drop a gene into the yellow field, and from the gene menu, you can select Get Sequence and get, for example, the peptide sequence for the gene as shown below:
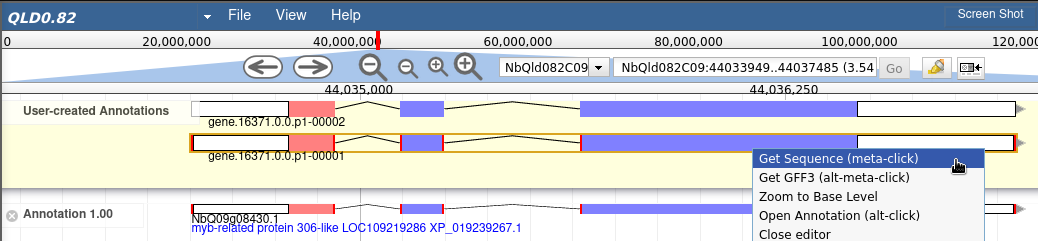
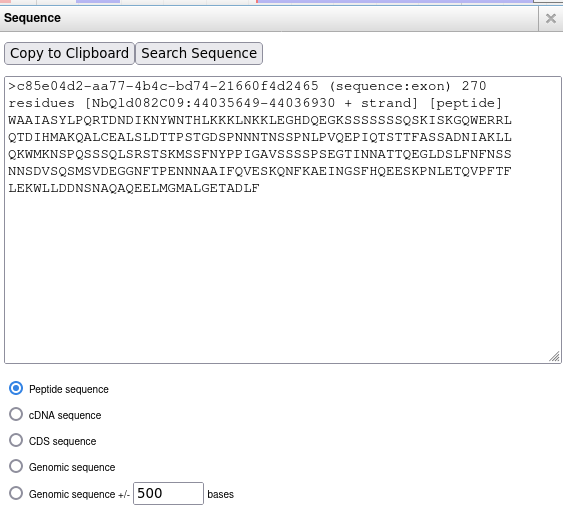
The above solution works, but most users forget to delete the genes from the yellow Apollo's annotation field. Therefore, I wrote a small web app based on Python's Flask framework, which can be found here. To add it to Apollo, I need to add the following lines to trackList.json for the gene track:
{
"menuTemplate": [
{},
{},
{
"action": "iframeDialog",
"label": "Get Sequence",
"title": "{Name}",
"iconClass": "dijitIconDatabase",
"url": "https://apollo.nbenth.com/getseq/{ID}"
}
],
"storeClass": "JBrowse/Store/SeqFeature/NCList",
"urlTemplate": "tracks/SeqViewer-test/{refseq}/trackData.jsonz",
"style": {
"className": "feature"
},
"label": "SeqViewer-test",
"apollo": {
"topType": "mRNA",
"source": "upload",
"type": "GFF3_JSON"
},
"type": "JBrowse/View/Track/HTMLFeatures",
"key": "SeqViewer-test"
}
"menuTemplate": [
{},
{},
{
"action": "iframeDialog",
"label": "Get Sequence",
"title": "{Name}",
"iconClass": "dijitIconDatabase",
"url": "https://apollo.nbenth.com/getseq/{ID}"
}
],
"storeClass": "JBrowse/Store/SeqFeature/NCList",
"urlTemplate": "tracks/SeqViewer-test/{refseq}/trackData.jsonz",
"style": {
"className": "feature"
},
"label": "SeqViewer-test",
"apollo": {
"topType": "mRNA",
"source": "upload",
"type": "GFF3_JSON"
},
"type": "JBrowse/View/Track/HTMLFeatures",
"key": "SeqViewer-test"
}
The user needs to select a gene and click right-mouse click and select Get Sequence from the menu:

However, the problem with my solution is that I need to create a new database for each annotation. How is it possible to access Apollo's database, or how can Get Sequence be extracted from Apollo and replace my code?
I really appreciate any help you can provide.
Michal
Michał T. Lorenc
Mar 9, 2022, 7:53:55 PM3/9/22
to apollo
P.S. I added it also to [GitHub issues](https://github.com/GMOD/Apollo/issues/2643)
Reply all
Reply to author
Forward
0 new messages
