File not found trackData.jsonz
9 views
Skip to first unread message
Michał T. Lorenc
Jul 13, 2022, 9:04:49 PM7/13/22
to apollo
Hi all,
I uploaded the following GFF3 file via Apollo's "New track" button:
$ ls -hlatr
-rw-r----- 1 debian debian 2.0G Jul 12 00:50 LabUsa16cWild01-20_C-N_snp.gff3
$ head LabUsa16cWild01-20_C-N_snp.gff3
##gff-version 3
NbLab360C00 SGSautoSNP SNP 121 121 . . . Name=NbLab3600000001;ID=NbLab3600000001;Contig name=NbLab360C00;SNP score=19;SNP pos. on scaffold=121;Genotype C=10*G;Genotype L=45*G;Genotype N=19*T;Genotype Q=23*G;Genotype S=82*G;Genotype T=18*G;Genotype U=54*G;Genotype W=24*G;Changes=G/T
NbLab360C00 SGSautoSNP SNP 409 409 . . . Name=NbLab3600000002;ID=NbLab3600000002;Contig name=NbLab360C00;SNP score=44;SNP pos. on scaffold=409;Genotype C=17*T;Genotype L=59*T;Genotype N=44*C;Genotype Q=54*T;Genotype S=106*T;Genotype T=41*T;Genotype U=95*T;Genotype W=51*T;Changes=C/T
NbLab360C00 SGSautoSNP SNP 519 519 . . . Name=NbLab3600000003;ID=NbLab3600000003;Contig name=NbLab360C00;SNP score=49;SNP pos. on scaffold=519;Genotype C=27*T;Genotype L=79*T;Genotype N=49*C;Genotype Q=63*T;Genotype S=105*T;Genotype T=55*T;Genotype U=99*T;Genotype W=77*T;Changes=C/T
##gff-version 3
NbLab360C00 SGSautoSNP SNP 121 121 . . . Name=NbLab3600000001;ID=NbLab3600000001;Contig name=NbLab360C00;SNP score=19;SNP pos. on scaffold=121;Genotype C=10*G;Genotype L=45*G;Genotype N=19*T;Genotype Q=23*G;Genotype S=82*G;Genotype T=18*G;Genotype U=54*G;Genotype W=24*G;Changes=G/T
NbLab360C00 SGSautoSNP SNP 409 409 . . . Name=NbLab3600000002;ID=NbLab3600000002;Contig name=NbLab360C00;SNP score=44;SNP pos. on scaffold=409;Genotype C=17*T;Genotype L=59*T;Genotype N=44*C;Genotype Q=54*T;Genotype S=106*T;Genotype T=41*T;Genotype U=95*T;Genotype W=51*T;Changes=C/T
NbLab360C00 SGSautoSNP SNP 519 519 . . . Name=NbLab3600000003;ID=NbLab3600000003;Contig name=NbLab360C00;SNP score=49;SNP pos. on scaffold=519;Genotype C=27*T;Genotype L=79*T;Genotype N=49*C;Genotype Q=63*T;Genotype S=105*T;Genotype T=55*T;Genotype U=99*T;Genotype W=77*T;Changes=C/T
However, the "16c vs LAB SNPs" worked fine but "16c vs NWA SNPs." does not load any content and the looks show this error:
2022-07-12 02:27:56,201 [http-nio-8080-exec-66] ERROR apollo.JbrowseController - File not found: /data/apollo_data/3135830-LAB360/tracks/16c_vs_NWA_SNPs/NbLab360C17/trackData.jsonz
Are there any file size restrictions for uploading via GUI?
Thank you in advance,
Michal
Nathan Dunn
Jul 14, 2022, 11:01:03 AM7/14/22
to "Michał T. Lorenc", apollo
There are upload limits. By default, they are pretty large (2 or 4 GBs?).
What “type” of track are you using (JSON I’m assuming?)?
It might be worth just trying to do it by hand with flatfile-to-json.pl from JBrowse to see if you get any errors for those tracks.
Nathan
--
To unsubscribe from this group and stop receiving emails from it, send an email to apollo+un...@lbl.gov.
Nathan Dunn
Jul 14, 2022, 11:07:18 PM7/14/22
to "Michał T. Lorenc", apollo
It was probably an error on processing the file.
Also, I’m not sure if it would know what to do with SNP types by default.
Conversely, I would just convert them to VCF files and upload them that way as I think the visualization would be more meaningful, though hopefully you have an existing VCF file you can pull from:
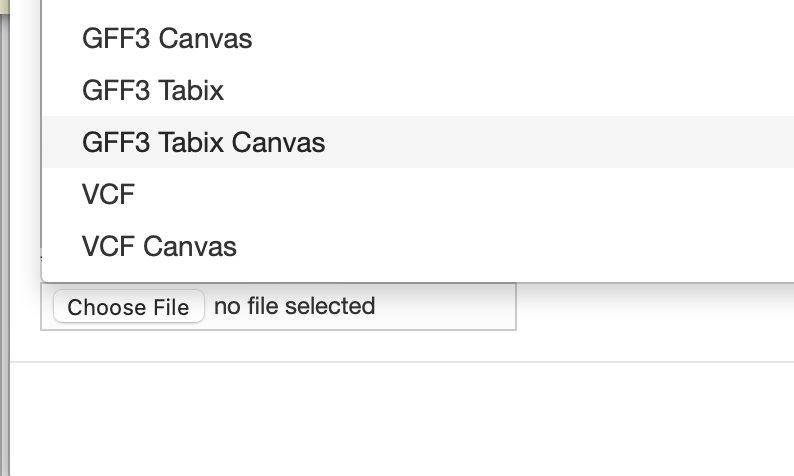
You could try updating the top type to “SNP”:
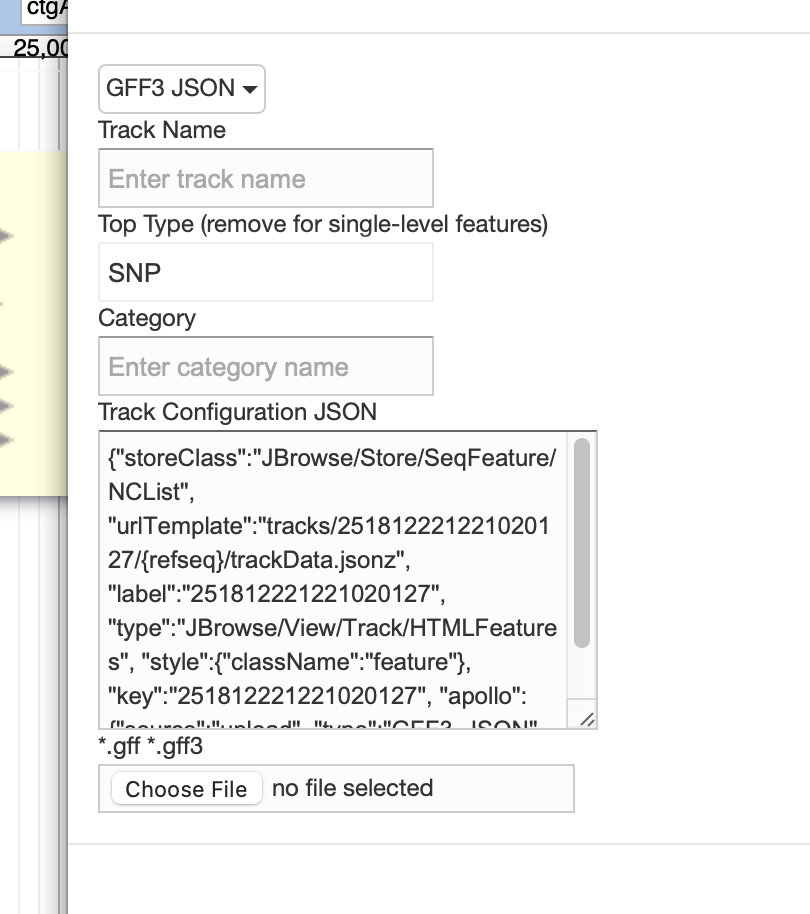
Nathan
On Jul 14, 2022, at 7:25 PM, Michał T. Lorenc <m.t.l...@gmail.com> wrote:Yes, Gff3 JSON.
Michał T. Lorenc
Jul 25, 2022, 10:34:36 PM7/25/22
to Nathan Dunn, apollo
Hi all,
It worked the whole time, but it took a long time. I solved the problem by compressing and indexing the files.
Best wishes,
Michal
Reply all
Reply to author
Forward
0 new messages
