HCP Early Psychosis Diffusion Data
Haley Wang
Glasser, Matt
Can you send a picture of the b0 image after HCP Pipelines preprocessing (i.e., in ${StudyFolder}/${Subject}/T1w/Diffusion/data.nii.gz?
Matt.
--
You received this message because you are subscribed to the Google Groups "HCP-Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
hcp-users+...@humanconnectome.org.
To view this discussion on the web visit
https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/c7f998e9-b5f2-46fb-98e5-18d4ec9841b2n%40humanconnectome.org.
The materials in this message are private and may contain Protected Healthcare Information or other information of a sensitive nature. If you are not the intended recipient, be advised that any unauthorized use, disclosure, copying or the taking of any action in reliance on the contents of this information is strictly prohibited. If you have received this email in error, please immediately notify the sender via telephone or return mail.
Haley Wang
Glasser, Matt
I would need to take a look at their diffusion MRI protocol PDF to comment further. Perhaps Jenn or Mike has this?
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Date: Wednesday, November 16, 2022 at 11:49 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Cc: "Glasser, Matt" <glas...@wustl.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
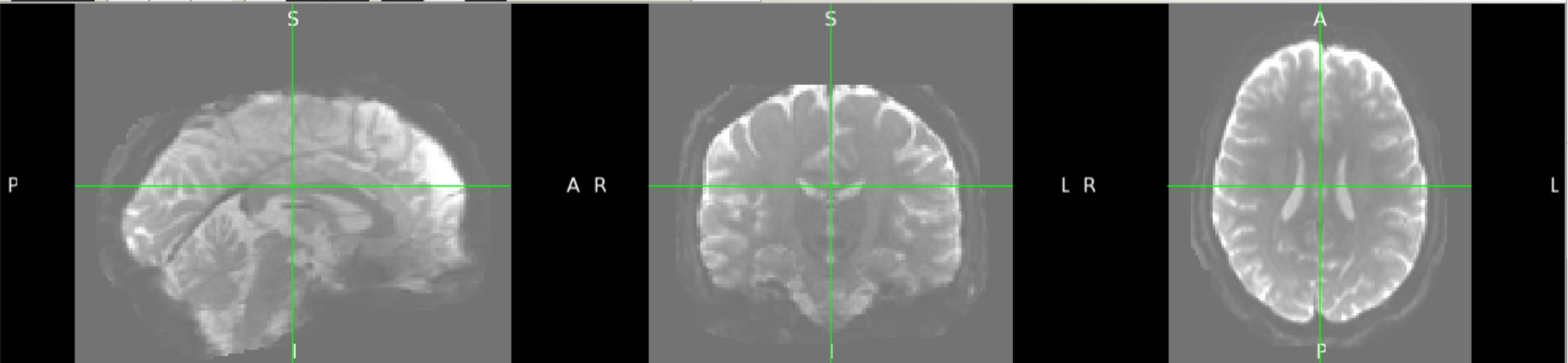
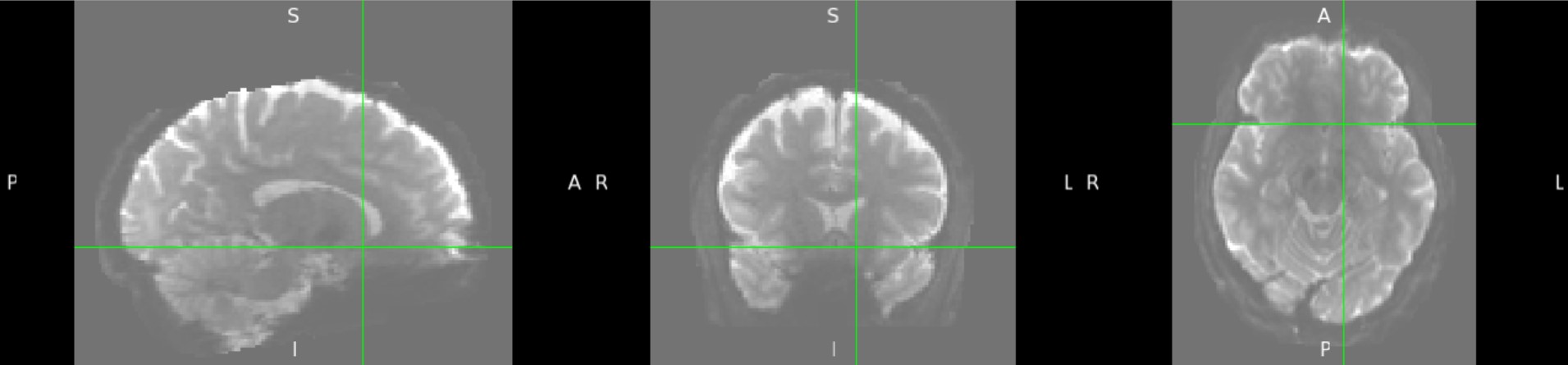
Yes - Here is subject 2029:


Haley Wang
Best,
Glasser, Matt
I’m not sure. I wasn’t involved in any of the CRHD projects.
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Date: Wednesday, November 16, 2022 at 11:54 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Cc: "Glasser, Matt" <glas...@wustl.edu>, "Elam, Jennifer" <el...@wustl.edu>, "Harms, Michael" <mha...@wustl.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
Thank you, Matt!
I also noticed that, different from other HCP datasets with b values of 1000, 2000, and 3000, HCP EP has 500, 1500, and 3000. Is there any reason for this choice?
Best,
Haley
On Wednesday, November 16, 2022 at 9:50:58 AM UTC-8 glas...@wustl.edu wrote:
I would need to take a look at their diffusion MRI protocol PDF to comment further. Perhaps Jenn or Mike has this?
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Date: Wednesday, November 16, 2022 at 11:49 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Cc: "Glasser, Matt" <glas...@wustl.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
Yes - Here is subject 2029:
On Wednesday, November 16, 2022 at 9:40:58 AM UTC-8 glas...@wustl.edu wrote:
Can you send a picture of the b0 image after HCP Pipelines preprocessing (i.e., in ${StudyFolder}/${Subject}/T1w/Diffusion/data.nii.gz?
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Reply-To: "hcp-...@humanconnectome.org" <hcp-...@humanconnectome.org>
Date: Wednesday, November 16, 2022 at 11:38 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Subject: [hcp-users] HCP Early Psychosis Diffusion Data
Hello HCP team,
I am working on preprocessing the diffusion images of the HCP EP 1.1 dataset, and I notice that more than half of the subjects have weak signals in the orbitofrontal and subcallosal cortex, even on raw b0 images. I used the HCP pipeline for preprocessing, but topup and eddy could only fix the distortion, not the signal strength. The eddy-processed image of about 90 subjects misses this area on their masks, due to extremely low signal, which makes it hard for later high-level analyses. I'm curious if this is a known issue for you, and if you have any suggestions on how to handle it. Thank you so much for your time and help!
All the best,
Haley
--
You received this message because you are subscribed to the Google Groups "HCP-Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to hcp-users+...@humanconnectome.org.
To view this discussion on the web visit https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/c7f998e9-b5f2-46fb-98e5-18d4ec9841b2n%40humanconnectome.org.
The materials in this message are private and may contain Protected Healthcare Information or other information of a sensitive nature. If you are not the intended recipient, be advised that any unauthorized use, disclosure, copying or the taking of any action in reliance on the contents of this information is strictly prohibited. If you have received this email in error, please immediately notify the sender via telephone or return mail.
The materials in this message are private and may contain Protected Healthcare Information or other information of a sensitive nature. If you are not the intended recipient, be advised that any unauthorized use, disclosure, copying or the taking of any action in reliance on the contents of this information is strictly prohibited. If you have received this email in error, please immediately notify the sender via telephone or return mail.
Elam, Jennifer
Sent: Wednesday, November 16, 2022 11:50 AM
To: Haley Wang <hale...@g.ucla.edu>; HCP-Users <hcp-...@humanconnectome.org>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
|
* External Email - Caution * |
Elam, Jennifer
Sent: Wednesday, November 16, 2022 12:13 PM
Glasser, Matt
With respect to the specific concerns about signal loss in regions of susceptibility:
The flip angles are 78 and 160 degrees, rather than 90 and 180. I don’t recall why that is, but it does potentially mean that we aren’t completely refocusing the spins in the areas with worst susceptibility.
Matt.
To view this discussion on the web visit https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/PH0PR02MB88284C47F04F6ACE033D5604CC079%40PH0PR02MB8828.namprd02.prod.outlook.com.
Harms, Michael
HCP-Lifespan protocol also used shells of b = 1500 and 3000.
HCP-YA is what used b = 1000, 2000, and 3000.
https://www.ncbi.nlm.nih.gov/pubmed/30261308
--
Michael Harms, Ph.D.
-----------------------------------------------------------
Associate Professor of Psychiatry
Washington University School of Medicine
Department of Psychiatry, Box 8134
660 South Euclid Ave. Tel: 314-747-6173
St. Louis, MO 63110 Email: mha...@wustl.edu
From: Haley Wang <hale...@g.ucla.edu>
Reply-To: "hcp-...@humanconnectome.org" <hcp-...@humanconnectome.org>
Date: Wednesday, November 16, 2022 at 11:54 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Cc: "Glasser, Matt" <glas...@wustl.edu>, "Elam, Jennifer" <el...@wustl.edu>, "Harms, Michael" <mha...@wustl.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
|
* External Email - Caution * |
Thank you, Matt!
I also noticed that, different from other HCP datasets with b values of 1000, 2000, and 3000, HCP EP has 500, 1500, and 3000. Is there any reason for this choice?
Best,
Haley
On Wednesday, November 16, 2022 at 9:50:58 AM UTC-8 glas...@wustl.edu wrote:
I would need to take a look at their diffusion MRI protocol PDF to comment further. Perhaps Jenn or Mike has this?
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Date: Wednesday, November 16, 2022 at 11:49 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Cc: "Glasser, Matt" <glas...@wustl.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
Yes - Here is subject 2029:
On Wednesday, November 16, 2022 at 9:40:58 AM UTC-8 glas...@wustl.edu wrote:
Can you send a picture of the b0 image after HCP Pipelines preprocessing (i.e., in ${StudyFolder}/${Subject}/T1w/Diffusion/data.nii.gz?
Matt.
From: Haley Wang <hale...@g.ucla.edu>
Reply-To: "hcp-...@humanconnectome.org" <hcp-...@humanconnectome.org>
Date: Wednesday, November 16, 2022 at 11:38 AM
To: HCP-Users <hcp-...@humanconnectome.org>
Subject: [hcp-users] HCP Early Psychosis Diffusion Data
Hello HCP team,
I am working on preprocessing the diffusion images of the HCP EP 1.1 dataset, and I notice that more than half of the subjects have weak signals in the orbitofrontal and subcallosal cortex, even on raw b0 images. I used the HCP pipeline for preprocessing, but topup and eddy could only fix the distortion, not the signal strength. The eddy-processed image of about 90 subjects misses this area on their masks, due to extremely low signal, which makes it hard for later high-level analyses. I'm curious if this is a known issue for you, and if you have any suggestions on how to handle it. Thank you so much for your time and help!
All the best,
Haley
--
You received this message because you are subscribed to the Google Groups "HCP-Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to hcp-users+...@humanconnectome.org.
To view this discussion on the web visit https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/c7f998e9-b5f2-46fb-98e5-18d4ec9841b2n%40humanconnectome.org.
The materials in this message are private and may contain Protected Healthcare Information or other information of a sensitive nature. If you are not the intended recipient, be advised that any unauthorized use, disclosure, copying or the taking of any action in reliance on the contents of this information is strictly prohibited. If you have received this email in error, please immediately notify the sender via telephone or return mail.
The materials in this message are private and may contain Protected Healthcare Information or other information of a sensitive nature. If you are not the intended recipient, be advised that any unauthorized use, disclosure, copying or the taking of any action in reliance on the contents of this information is strictly prohibited. If you have received this email in error, please immediately notify the sender via telephone or return mail.
--
You received this message because you are subscribed to the Google Groups "HCP-Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
hcp-users+...@humanconnectome.org.
To view this discussion on the web visit https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/da22695d-783f-429f-bc71-2835f624c229n%40humanconnectome.org.
Harms, Michael
The dMRI acquisitions for HCP-EP appear to be the same as for HCP-Lifespan (except they collected some volumes with b=500 as well).
The flip angles of 78/160 are what we used for HCP-EP as well. We intentionally “underflip” a little bit relative to the traditional 90/180, since the center of the brain tends to be “overflipped” if you call for 90/180.
I don’t see any reason that the HCP-EP dMRI data should be different from the HCP-Lifespan data.
Cheers,
-MH
--
Michael Harms, Ph.D.
-----------------------------------------------------------
Associate Professor of Psychiatry
Washington University School of Medicine
Department of Psychiatry, Box 8134
660 South Euclid Ave. Tel: 314-747-6173
St. Louis, MO 63110 Email: mha...@wustl.edu
From: "Glasser, Matt" <glas...@wustl.edu>
Reply-To: "hcp-...@humanconnectome.org" <hcp-...@humanconnectome.org>
Date: Wednesday, November 16, 2022 at 2:22 PM
To: "hcp-...@humanconnectome.org" <hcp-...@humanconnectome.org>, Haley Wang <hale...@g.ucla.edu>, "sylvai...@etsmtl.ca" <sylvai...@etsmtl.ca>, Mike Coleman <m...@bwh.harvard.edu>
Subject: Re: [hcp-users] HCP Early Psychosis Diffusion Data
|
* External Email - Caution * |
To view this discussion on the web visit https://groups.google.com/a/humanconnectome.org/d/msgid/hcp-users/60E6C374-CFD7-4D46-B672-BE475181F651%40wustl.edu.
